Using RStudio
Warning
Using RStudio differs between different HPC clusters.
We also recommend ThinLinc!
On UPPMAX
There is a system installed version, available via the “rstudio” command (you will get RStudio/1.1.423).
However, we recommend you to use a RStudio module.
Warning
We recommend ThinLinc because the graphics is more effective there!
Using ThinLinc
ThinLinc app:
<user>@rackham-gui.uppmax.uu.seThinLinc in web browser:
https://rackham-gui.uppmax.uu.seThis requires 2FA!Choose Xfce as the desktop environment (faster)
start a command line window
Using terminal
Remember to have X11 installed!
On Mac
install XQuartz
On Windows
Use MobaXterm or
Check for RStudio versions
Check all available versions with:
$ module avail RStudio
Output at UPPMAX as of March 10 2024
[bjornc@rackham5 ~]$ ml av RStudio ------------------------------------- /sw/mf/rackham/applications ------------------------------------- RStudio/1.0.136 RStudio/1.1.423 RStudio/2022.02.0-443 RStudio/2023.06.0-421 RStudio/1.0.143 RStudio/1.1.463 RStudio/2022.02.3-492 RStudio/2023.06.2-561 RStudio/1.0.153 RStudio/1.4.1106 RStudio/2022.07.1-554 RStudio/2023.12.1-402 (D) Where: D: Default Module Use "module spider" to find all possible modules and extensions. Use "module keyword key1 key2 ..." to search for all possible modules matching any of the "keys".
load R_packages
module load RStudio
run
rstudio &from the command line, and waitit might take 5-10 minutes for RStudio to start, especially if you loaded R_packages as well, but once it starts, there should be no further delays
do not start RStudio through the graphical menu system in ThinLinc, this will not have access to loaded modules.
if it takes a long time and might be due to that you have saved a lot of workspace
Example:
Demo
$ module load R/4.1.1
$ module load RStudio/2023.12.1-402
$ rstudio &
If you’re going to run heavier computations within RStudio then you have to remember that you need to do it inside an interactive session on one of the computation nodes, and not on a login node. But if you mostly want to use it as a pretty code editor then you can run it on the login node as well.
To use Rstudio on a compute node, start by asking SLURM for an interactive allocation (within the ThinLink session). E.g.
$ interactive -A naiss2023-22-44 -p devcore -n 4 -t 10:00
On Bianca
When logging onto Bianca, you are placed on a login node, which nowadays has 2 CPU and a few GB of RAM. This is sufficient for doing some light-weight calculations, but interactive sessions and batch jobs provide access to much more resources and should be requested via the SLURM system.
The desktop client version of ThinLinc does not work for Bianca. Instead you run and login to ThinLinc in the browser:
On HPC2N
Rstudio also exists on Kebnekaise but is only installed on the ThinLinc login nodes and not on the compute nodes (and also not on the regular login nodes accessible with SSH). Thus, Rstudio should only be used for development and very light analysis, since there is no way to submit a job to the compute nodes.
Login to ThinLinc desktop application by providing the following
server: kebnekaise-tl.hpc2n.umu.se
username
password
Alternatively, you can use ThinLinc in the browser: https://kebnekaise-tl.hpc2n.umu.se:300/
When in ThinLinc, you can start Rstudio either from the menu (version 4.0.4) or from the command line. If you start it from the command line you first need to load R and its prerequisites, but you can pick between several versions this way.
At LUNARC
At LUNARC the recommended way to run RStudio is in Thinlinc with Desktop On Demand. All available versions are in the Applications menu under Applications-R.
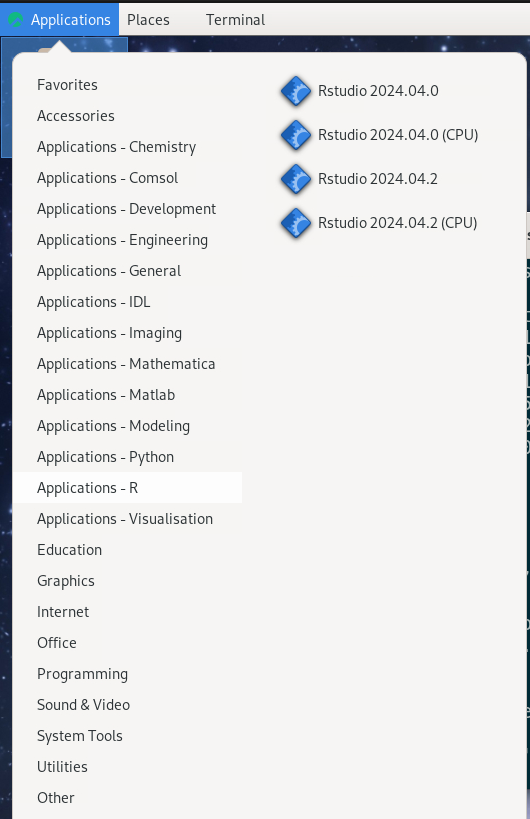
There is a regular version and a “(CPU)” version for each release. - Regular versions run an Intel 32-core node with a GPU partition, but the wall time limit is 48 hours. - CPU versions run on an AMD 48-core CPU-only node, and allows users to set a wall time of up to 168 hours (7 days), at the cost of reduced graphical support.
There is no need to pre-load any modules before starting RStudio from the Desktop On Demand Applications-R menu.
If you run from the command line, you will need to load R/4.4.1 (check prerequisite versions of GCC and OpenMPI with ml spider R/4.4.1) and one of the RStudio modules above. Please only launch RStudio from one of the Desktop On Demand terminals, not the front-end terminal in the Favorites menu.
ml avail output at LUNARC as of October 21 2024
[<user>@cosmos2 /]$ ml avail RStudio --------------------- /sw/easybuild_milan/modules/all/Core --------------------- rstudio/2024.04.0-735 rstudio/2024.04.2-764 (D) Where: D: Default Module If the avail list is too long consider trying: "module --default avail" or "ml -d av" to just list the default modules. "module overview" or "ml ov" to display the number of modules for each name. Use "module spider" to find all possible modules and extensions. Use "module keyword key1 key2 ..." to search for all possible modules matching any of the "keys".