Parallel and multithreaded functions
Questions
What is parallel programming?
Why do we need it?
When can I use it?
Objectives
Learn basic concepts in parallel programming
Gain knowledge on the tools for parallel programming in different languages
Familiarize with the tools to monitor the usage of resources
What is parallel programming?
Parallel programming is the science and art of writing code that execute tasks on different computing units (cores) simultaneously. In the past computers were shiped with a single core per Central Processing Unit (CPU) and therefore only a single computation at the time (serial program) could be executed.
Nowadays computer architectures are more complex than the single core CPU mentioned already. For instance, common architectures include those where several cores in a CPU share a common memory space and also those where CPUs are connected through some network interconnect.
Shared Memory and Distributed Memory architectures.
A more realistic picture of a computer architecture can be seen in the following picture where we have 14 cores that shared a common memory of 64 GB. These cores form the socket and the two sockets shown in this picture constitute a node.
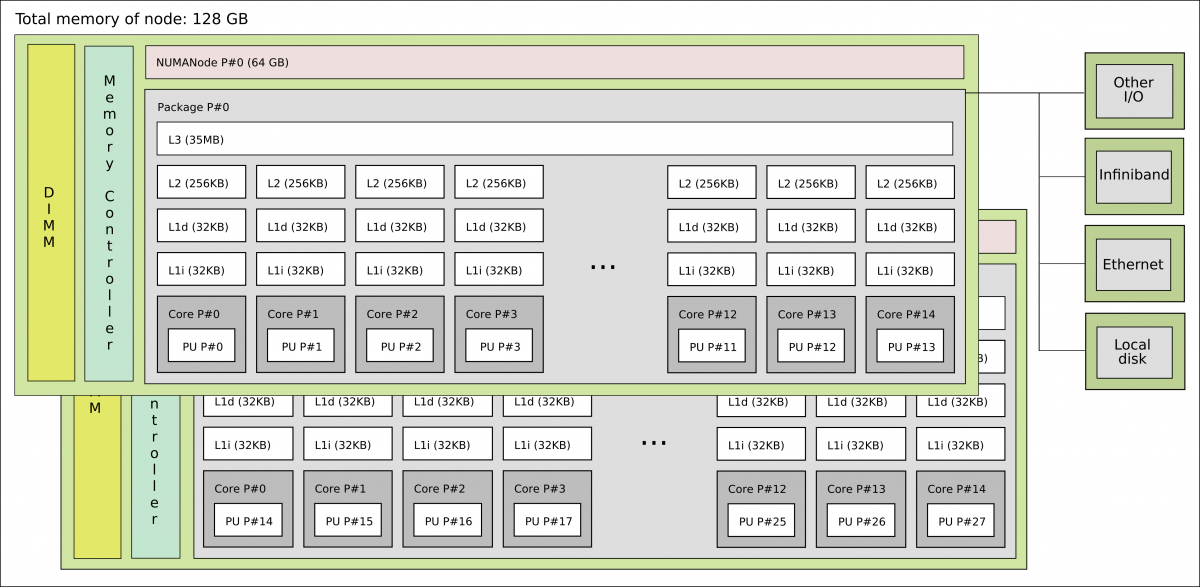
1 standard node on Kebnekaise @HPC2N
It is interesting to notice that there are different types of memory available for the cores, ranging from the L1 cache to the node’s memory for a single node. In the former, the bandwidth can be TB/s while in the latter GB/s.
Now you can see that on a single node you already have several computing units (cores) and also a hierarchy of memory resources which is denoted as Non Uniform Memory Access (NUMA).
Besides the standard CPUs, nowadays one finds Graphic Processing Units (GPUs) architectures in HPC clusters.
Why is parallel programming needed?
There is no “free lunch” when trying to use features (computing/memory resources) in modern architectures. If you want your code to be aware of those features, you will need to either add them explicitly (by coding them yourself) or implicitly (by using libraries that were coded by others).
In your local machine, you may have some number of cores available and some memory attached to them which can be exploited by using a parallel program. There can be some limited resources for running your data-production simulations as you may use your local machine for other purposes such as writing a manuscript, making a presentation, etc. One alternative to your local machine can be a High Performance Computing (HPC) cluster another could be a cloud service. A common layout for the resources in an HPC cluster is a shown in the figure below.
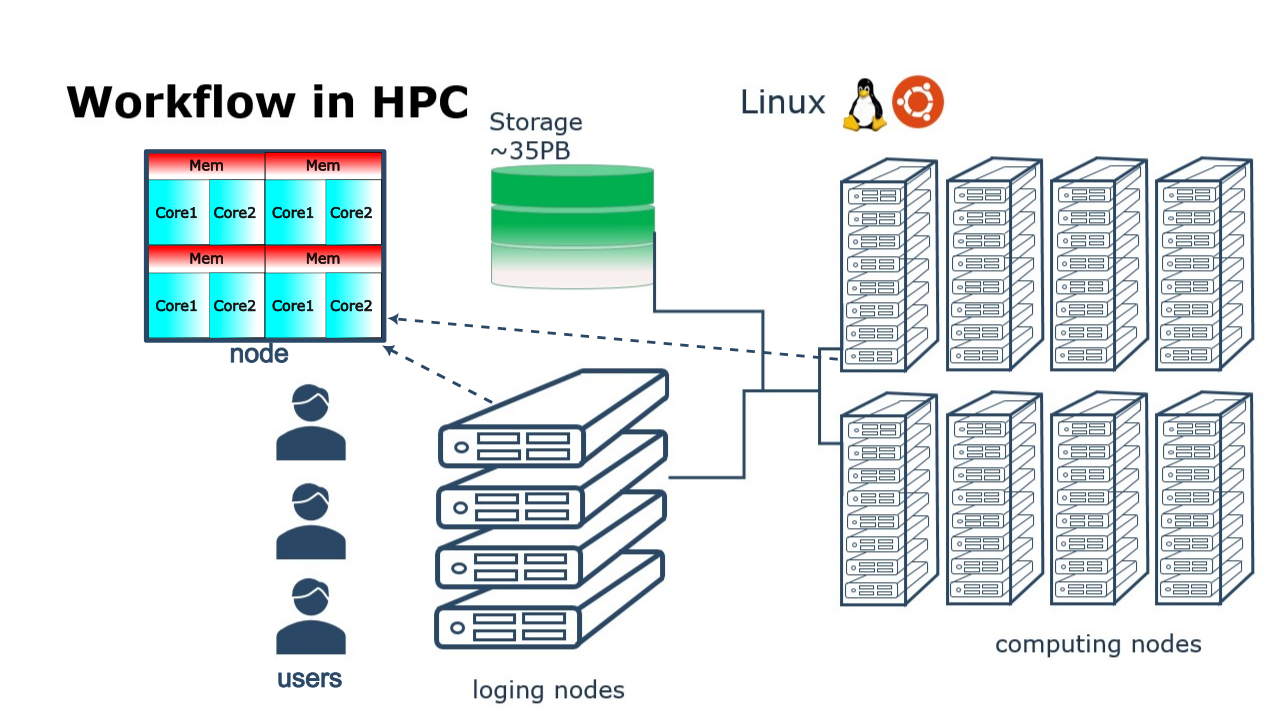
High Performance Computing (HPC) cluster.
Although a serial application can run in such a cluster, it would not gain much of the HPC resources. If fact, one can underuse the cluster if one allocates more resources than what the simulation requires.
Under-using a cluster.
Warning
Check if the resources that you allocated are being used properly.
Monitor the usage of hardware resources with tools offered at your HPC center, for instance job-usage at HPC2N.
Here there are some examples (of many) of what you will need to pay attention when porting a parallel code from your laptop (or another HPC center) to our clusters:
We have a tool to monitor the usage of resources called: job-usage at HPC2N.
If you are in a interactive node session the top command will give you information
of the resources usage.
Common parallel programming paradigms
Now the question is how to take advantage of modern architectures which consist of many-cores, interconnected through networks, and that have different types of memory available? Python, Julia, Matlab, and R languages have different tools and libraries that can help you to get more from your local machine or HPC cluster resources.
Threaded programming
To take advantage of the shared memory of the cores, threaded mechanisms can be used.
Low-level programming languages, such as Fortran/C/C++, use OpenMP as the standard
application programming interface (API) to parallelize programs by using a threaded mechanism.
Here, all threads have access to the same data and can do computations simultaneously.
From this we infer that without doing any modification to our code
we can get the benefits from parallel computing by turning-on/off external libraries,
by setting environment variables such as OMP_NUM_THREADS.
Higher-level languages have their own mechanisms to generate threads and this can be confusing especially if the code is using external libraries, linear algebra for instance (LAPACK, BLAS, …). These libraries have their own threads (OpenMP for example) and the code you are writing (R, Julia, Python, or Matlab) can also have some internal threded mechanism.
Warning
Check if the libraries/packages that you are using have a threaded mechanism.
Monitor the usage of hardware resources with tools offered at your HPC center, for instance job-usage at HPC2N.
Here there are some examples (of many) of what you will need to pay attention when porting a parallel code from your laptop (or another HPC center) to our clusters:
For some linear algebra operations Numpy supports threads (set with the OMP_NUM_THREADS variable).
If your code contains calls to these operations in a loop that is already parallelized by n processes,
and you allocate n cores for this job, this job will exceed the allocated resources unless the
number of threads is explicitly set to 1.
For some linear algebra operations Julia supports threads (set with the OMP_NUM_THREADS variable).
If your code contains calls to these operations in a loop that is already parallelized by n processes,
and you allocate n cores for this job, this job will exceed the allocated resources unless the
number of threads is explicitly set to 1. Notice that Julia also has its own threaded mechanism.
Creating a cluster with n cores (makeCluster) and start traing a ML model with flags such as
allowParallel set to TRUE or num.threads set to a value such as the total number of requested
cores is exceeded.
Using a CPLEX solver inside a parfor loop: These solvers work in a opportunistic manner meaning that
they will try to use all the resources available in the machine. If you request n cores for parfor in
your batch job, these cores will be used by the solver. Theoretically, you will be using nxn cores although
only n were requested. One way to solve this issue is by setting the number of threads
cplex.Param.threads.Cur to 1.
A common issue with shared memory programming is data racing which happens when different threads write on the same memory address.
Language-specific nuances for threaded programming
Python offers its own threaded mechanism but due to a locking mechanism, Python threads are not efficient for computation. However, Python threads could be useful for I/O files handling. Code modifications are required to support the threads.
The mechanism here is called Julia threads which is performant and can be activated by
executing a script as follows julia --threads X script.jl, where X is the number of
threads. Code modifications are required to support the threads.
R doesn’t have a threaded mechanism as the other languages discussed in this course. Some functions provided by certain packages (parallel, doParallel, etc.), for instance, foreach, offer parallel features but memory is not shared across the workers. This could lead to data replication.
Starting from version 2020a, Matlab offers the ThreadPool functionality that can leverage the power of threads sharing a common memory. This could potentially lead to a faster code compared to other schemes (Distributed discussed below) but notice that the code is not expected to support multi-node simulations.
Distributed programming
Although threaded programming is convenient because one can achieve considerable initial speedups with little code modifications, this approach does not scale for more than hundreds of cores. Scalability can be achieved with distributed programming. Here, there is not a common shared memory but the individual processes (notice the different terminology with threads in shared memory) have their own memory space. Then, if a process requires data from or should transfer data to another process, it can do that by using send and receive to transfer messages. A standard API for distributed computing is the Message Passing Interface (MPI). In general, MPI requires refactoring of your code.
Language-specific nuances for distributed programming
Python has different modules for achieving distributed programming, for instance multiprocessing and
mpi4py. The former is part of the Python standard library so you don’t need to do further installations,
while the latter needs to be installed. Also, one needs to learn the concepts of MPI prior to using the
feautures offered by this module.
The mechanism here is called Julia processes which can be activated by executing a script as follows
julia -p X script.jl, where X is the number of processes. Code modifications are required to support the
workers. Julia also supports MPI through the package MPI.jl.
R doesn’t have a multiprocessing mechanism as the other languages discussed in this course. Some
functions provided by certain packages (parallel, doParallel, etc.), for instance, foreach,
offer parallel features. The processes generated by these functions have their own workspace which
could lead to data replication.
MPI is supported in R through the Rmpi package.
In Matlab one can use the parpool('my-cluster',X) where X is the number of workers. The total number of processes spawned will always be X+1 where the extra process handles the overhead for the rest. See the
documentation for parpool from MatWorks.
Matlab doesn’t support MPI function calls in Matlab code, it could be used indirectly through
mex functions though.
Big data
Sometimes the workflow you are targeting doesn’t require extensive computations but mainly dealing with big pieces of data. An example can be, reading a column-structured file and doing some transformation per-column. Fortunately, all languages covered in this course have already several tools to deal with big data. We list some of these tools in what follows but notice that other tools doing similar jobs can be available for each language.
Language-specific tools for big data
Dask
Dask is a array model extension and task scheduler. By using the new array classes, you can automatically distribute operations across multiple CPUs.
Dask is very popular for data analysis and is used by a number of high-level Python libraries:
Dask arrays scale NumPy (see also xarray)
Dask dataframes scale Pandas workflows
Dask-ML scales Scikit-Learn
Dask divides arrays into many small pieces (chunks), as small as necessary to fit it into memory.
Operations are delayed (lazy computing) e.g. tasks are queue and no computation is performed until you actually ask values to be computed (for instance print mean values).
Then data is loaded into memory and computation proceeds in a streaming fashion, block-by-block.
An example of a Jupyter notebook running Dask can be found here.
Dagger
According to the developers of this framework, Dagger is heavily inspired on Dask. It support distributed arrays so that they could fit the memory and also the possibility of parallelizing the computations on these arrays.
Arrow (previously disk.frame) can deal with big arrays. Other tools include data.table and bigmemory.
In Matlab Tall Arrays and Distributed Arrays will assist you when dealing with large arrays.
Demo
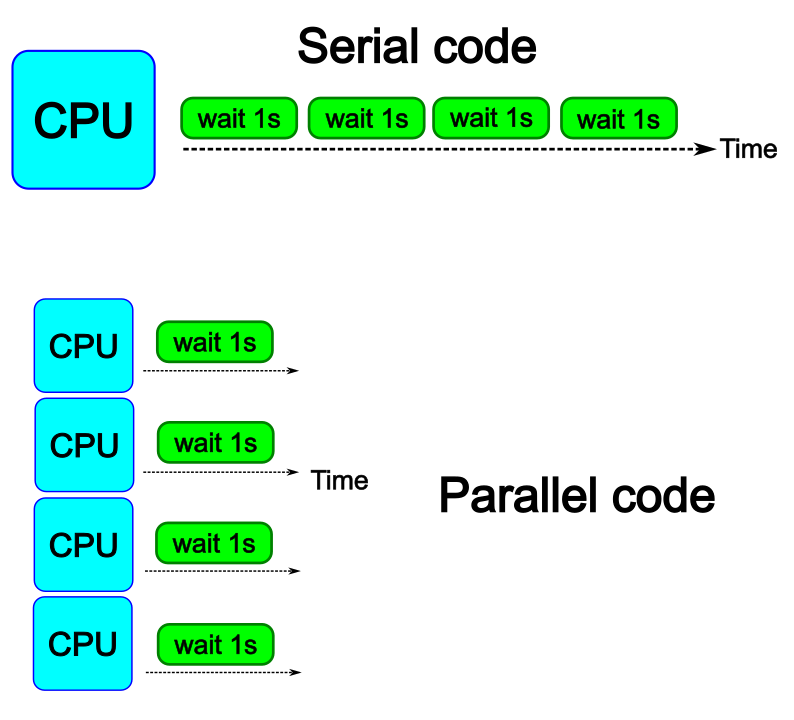
The idea is to parallelize a simple for loop (language-agnostic):
for i start at 1 end at 4
wait 1 second
end the for loop
The waiting step is used to simulate a task without writing too much code. In this way, one can realize how faster the loop can be executed when threads are added:
In the following example sleep.py the sleep() function is called n times first in
serial mode and then by using n processes. To parallelize the serial code we can use
the multiprocessing module that is shipped with the base library in Python so that
you don’t need to install it.
import sys
from time import perf_counter,sleep
import multiprocessing
# number of iterations
n = 4
# number of processes
numprocesses = 4
def sleep_serial(n):
for i in range(n):
sleep(1)
def sleep_threaded(n,numprocesses,processindex):
# workload for each process
workload = n/numprocesses
begin = int(workload*processindex)
end = int(workload*(processindex+1))
for i in range(begin,end):
sleep(1)
if __name__ == "__main__":
starttime = perf_counter() # Start timing serial code
sleep_serial(n)
endtime = perf_counter()
print("Time spent serial: %.2f sec" % (endtime-starttime))
starttime = perf_counter() # Start timing parallel code
processes = []
for i in range(numprocesses):
p = multiprocessing.Process(target=sleep_threaded, args=(n,numprocesses,i))
processes.append(p)
p.start()
# waiting for the processes
for p in processes:
p.join()
endtime = perf_counter()
print("Time spent parallel: %.2f sec" % (endtime-starttime))
First load the modules ml GCCcore/11.2.0 Python/3.9.6 and then run the script
with the command srun -A "your-project" -n 1 -c 4 -t 00:05:00 python sleep.py to use 4 processes.
In the following example sleep-threads.jl the sleep() function is called n times
first in serial mode and then by using n threads. The BenchmarkTools package
help us to time the code (as this package is not in the base Julia installation you will need
to install it).
using BenchmarkTools
using .Threads
n = 4 # number of iterations
function sleep_serial(n) #Serial version
for i in 1:n
sleep(1)
end
end
@btime sleep_serial(n) evals=1 samples=1
function sleep_threaded(n) #Parallel version
@threads for i = 1:n
sleep(1)
end
end
@btime sleep_threaded(n) evals=1 samples=1
First load the Julia module ml Julia/1.8.5-linux-x86_64 and then run the script
with the command srun -A "your-project" -n 1 -c 4 -t 00:05:00 julia --threads 4 sleep-threads.jl
to use 4 Julia threads.
We can also use the Distributed package that allows the scaling of simulations beyond
a single node (call the script sleep-distributed.jl):
using BenchmarkTools
using Distributed
n = 4 # number of iterations
function sleep_parallel(n)
@sync @distributed for i in 1:n
sleep(1)
end
end
@btime sleep_parallel(n) evals=1 samples=1
Run the script with the command srun -A "your-project" -n 1 -c 4 -t 00:05:00 julia -p 4 sleep-distributed.jl
to use 4 Julia processes.
In the following example sleep.R the Sys.sleep() function is called n times
first in serial mode and then by using n processes. Start by loading the
modules ml GCC/12.2.0 OpenMPI/4.1.4 R/4.2.2
library(doParallel)
# number of iterations = number of processes
n <- 4
sleep_serial <- function(n) {
for (i in 1:n) {
Sys.sleep(1)
}
}
serial_time <- system.time( sleep_serial(n) )[3]
serial_time
sleep_parallel <- function(n) {
r <- foreach(i=1:n) %dopar% Sys.sleep(1)
}
cl <- makeCluster(n)
registerDoParallel(cl)
parallel_time <- system.time( sleep_parallel(n) )[3]
stopCluster(cl)
parallel_time
Run the script with the command srun -A "your-project" -n 1 -c 4 -t 00:05:00 Rscript --no-save --no-restore sleep.R.
In Matlab one can use the function pause() to wait for some number of secods.
The Matlab module we tested can be loaded as ml MATLAB/2023a.Update4.
% Get a handler for the cluster
c=parcluster('kebnekaise');
n = 4; % Number of iterations
% Run the job with 1 worker and submit the job to the batch queue
j = c.batch(@sleep_serial, 1, {4}, 'pool', 1);
% Wait till the job has finished
j.wait;
% Fetch the result after the job has finished
t = j.fetchOutputs{:};
fprintf('Time taken for serial version: %.2f seconds\n', t);
% Run the job with 4 worker and submit the job to the batch queue
j = c.batch(@sleep_parallel, 1, {4}, 'pool', 4);
% Wait till the job has finished
j.wait;
% Fetch the result after the job has finished
t = j.fetchOutputs{:};
fprintf('Time taken for parallel version: %.2f seconds\n', t);
% Serial version
function t_serial = sleep_serial(n)
% Start timming
tic;
for i = 1:n
pause(1);
end
t_serial = toc; % stop timing
end
% Parallel version
function t_parallel = sleep_parallel(n)
% Start timing
tic;
parfor i = 1:n
pause(1);
end
t_parallel = toc; % stop timing
end
You can run this code directly in the Matlab GUI.
Exercises
Running a parallel code efficiently
In this exercise we will run a parallelized code that performs a 2D integration:
\[\int^{\pi}_{0}\int^{\pi}_{0}\sin(x+y)dxdy = 0\]
One way to perform the integration is by creating a grid in the x and y directions.
More specifically, one divides the integration range in both directions into n bins.
Here is a parallel code using the multiprocessing module in Python (call it
integration2d_multiprocessing.py):
integration2d_multiprocessing.py
import multiprocessing
from multiprocessing import Array
import math
import sys
from time import perf_counter
# grid size
n = 5000
# number of processes
numprocesses = *FIXME*
# partial sum for each thread
partial_integrals = Array('d',[0]*numprocesses, lock=False)
# Implementation of the 2D integration function (non-optimal implementation)
def integration2d_multiprocessing(n,numprocesses,processindex):
global partial_integrals;
# interval size (same for X and Y)
h = math.pi / float(n)
# cummulative variable
mysum = 0.0
# workload for each process
workload = n/numprocesses
begin = int(workload*processindex)
end = int(workload*(processindex+1))
# regular integration in the X axis
for i in range(begin,end):
x = h * (i + 0.5)
# regular integration in the Y axis
for j in range(n):
y = h * (j + 0.5)
mysum += math.sin(x + y)
partial_integrals[processindex] = h**2 * mysum
if __name__ == "__main__":
starttime = perf_counter()
processes = []
for i in range(numprocesses):
p = multiprocessing.Process(target=integration2d_multiprocessing, args=(n,numprocesses,i))
processes.append(p)
p.start()
# waiting for the processes
for p in processes:
p.join()
integral = sum(partial_integrals)
endtime = perf_counter()
print("Integral value is %e, Error is %e" % (integral, abs(integral - 0.0)))
print("Time spent: %.2f sec" % (endtime-starttime))
Run the code with the following batch script.
job.sh
#!/bin/bash -l
#SBATCH -A naiss202X-XY-XYZ # your project_ID
#SBATCH -J job-serial # name of the job
#SBATCH -n *FIXME* # nr. tasks/coresw
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
# Load any modules you need, here for Python 3.11.8 and compatible SciPy-bundle
module load python/3.11.8
python integration2d_multiprocessing.py
#!/bin/bash
#SBATCH -A hpc2n202X-XYZ # your project_ID
#SBATCH -J job-serial # name of the job
#SBATCH -n *FIXME* # nr. tasks
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
# Do a purge and load any modules you need, here for Python
ml purge > /dev/null 2>&1
ml GCCcore/11.2.0 Python/3.9.6
python integration2d_multiprocessing.py
#!/bin/bash
#SBATCH -A lu202X-XX-XX # your project_ID
#SBATCH -J job-serial # name of the job
#SBATCH -n *FIXME* # nr. tasks
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
# reservation (optional)
#SBATCH --reservation=RPJM-course*FIXME*
# Do a purge and load any modules you need, here for Python
ml purge > /dev/null 2>&1
ml GCCcore/12.3.0 Python/3.11.3
python integration2d_multiprocessing.py
Try different number of cores for this batch script (FIXME string) using the sequence: 1,2,4,8,12, and 14. Note: this number should match the number of processes (also a FIXME string) in the Python script. Collect the timings that are printed out in the job.*.out. According to these execution times what would be the number of cores that gives the optimal (fastest) simulation?
Challenge: Increase the grid size (n) to 15000 and submit the batch job with 4 workers (in the
Python script) and request 5 cores in the batch script. Monitor the usage of resources
with tools available at your center, for instance top (UPPMAX) or
job-usage (HPC2N).
Here is a parallel code using the Distributed package in Julia (call it
integration2d_distributed.jl):
integration2D_distributed.jl
using Distributed
using SharedArrays
using LinearAlgebra
using Printf
using Dates
# Add worker processes (replace with actual number of cores you want to use)
nworkers = *FIXME*
addprocs(nworkers)
# Grid size
n = 20000
# Number of processes
numprocesses = nworkers
# Shared array to store partial sums for each process
partial_integrals = SharedVector{Float64}(numprocesses)
# Function for 2D integration using multiprocessing
# the decorator @everywher instruct Julia to transfer this function to all workers
@everywhere function integration2d_multiprocessing(n, numprocesses, processindex, partial_integrals)
# Interval size (same for X and Y)
h = π / n
# Cumulative variable
mysum = 0.0
# Workload for each process
workload = div(n, numprocesses)
# Define the range of work for each process according to index
begin_index = workload * (processindex - 1) + 1
end_index = workload * processindex
# Regular integration in the X axis
for i in begin_index:end_index
x = h * (i - 0.5)
# Regular integration in the Y axis
for j in 1:n
y = h * (j - 0.5)
mysum += sin(x + y)
end
end
# Store the result in the shared array
partial_integrals[processindex] = h^2 * mysum
end
# function for main
function main()
# Start the timer
starttime = now()
# Distribute tasks to processes
@sync for i in 1:numprocesses
@spawnat i integration2d_multiprocessing(n, numprocesses, i, partial_integrals)
end
# Calculate the total integral by summing over partial integrals
integral = sum(partial_integrals)
# end timing
endtime = now()
# Output results
println("Integral value is $(integral), Error is $(abs(integral - 0.0))")
println("Time spent: $(Dates.value(endtime - starttime) / 1000) sec")
end
# Run the main function
main()
Run the code with the following batch script.
job.sh
#!/bin/bash -l
#SBATCH -A naiss202X-XY-XYZ # your project_ID
#SBATCH -J job-serial # name of the job
#SBATCH -n *FIXME* # nr. tasks/coresw
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
ml julia/1.8.5
julia integration2D_distributed.jl
#!/bin/bash
#SBATCH -A hpc2n202x-xyz # your project_ID
#SBATCH -J job-serial # name of the job
#SBATCH -n *FIXME* # nr. tasks
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
ml purge > /dev/null 2>&1
ml Julia/1.9.3-linux-x86_64
julia integration2D_distributed.jl
#!/bin/bash
#SBATCH -A lu202X-XX-XX # your project_ID
#SBATCH -J job-serial # name of the job
#SBATCH -n *FIXME* # nr. tasks
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
# reservation (optional)
#SBATCH --reservation=RPJM-course*FIXME*
ml purge > /dev/null 2>&1
ml Julia/1.9.3-linux-x86_64
julia integration2D_distributed.jl
Try different number of cores for this batch script (FIXME string) using the sequence: 1,2,4,8,12, and 14. Note: this number should match the number of processes (also a FIXME string) in the Julia script. Collect the timings that are printed out in the job.*.out. According to these execution times what would be the number of cores that gives the optimal (fastest) simulation?
Challenge: Increase the grid size (n) to 100000 and submit the batch job with 4 workers (in the
Julia script) and request 5 cores in the batch script. Monitor the usage of resources
with tools available at your center, for instance top (UPPMAX) or
job-usage (HPC2N).
Here is a parallel code using the parallel and doParallel packages in R (call it
integration2d.R). Note: check if those packages are already installed for the required
R version, otherwise install them with install.packages(). The recommended R version
for this exercise is ml GCC/12.2.0 OpenMPI/4.1.4 R/4.2.2 (HPC2N).
integrationd.R
library(parallel)
library(doParallel)
# nr. of workers/cores that will solve the tasks
nworkers <- *FIXME*
# grid size
n <- 840
# Function for 2D integration (non-optimal implementation)
integration2d <- function(n, numprocesses, processindex) {
# Interval size (same for X and Y)
h <- pi / n
# Cumulative variable
mysum <- 0.0
# Workload for each process
workload <- floor(n / numprocesses)
# Define the range of work for each process according to index
begin_index <- workload * (processindex - 1) + 1
end_index <- workload * processindex
# Regular integration in the X axis
for (i in begin_index:end_index) {
x <- h * (i - 0.5)
# Regular integration in the Y axis
for (j in 1:n) {
y <- h * (j - 0.5)
mysum <- mysum + sin(x + y)
}
}
# Return the result
return(h^2 * mysum)
}
# Set up the cluster for doParallel
cl <- makeCluster(nworkers)
registerDoParallel(cl)
# Start the timer
starttime <- Sys.time()
# Distribute tasks to processes and combine the outputs into the results list
results <- foreach(i = 1:nworkers, .combine = c) %dopar% { integration2d(n, nworkers, i) }
# Calculate the total integral by summing over partial integrals
integral <- sum(results)
# End the timing
endtime <- Sys.time()
# Print out the result
print(paste("Integral value is", integral, "Error is", abs(integral - 0.0)))
print(paste("Time spent:", difftime(endtime, starttime, units = "secs"), "seconds"))
# Stop the cluster after computation
stopCluster(cl)
Run the code with the following batch script.
job.sh
#!/bin/bash -l
#SBATCH -A naiss202X-XY-XYZ # your project_ID
#SBATCH -J job-serial # name of the job
#SBATCH -n *FIXME* # nr. tasks/coresw
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
ml R_packages/4.1.1
Rscript --no-save --no-restore integration2d.R
#!/bin/bash
#SBATCH -A hpc2n202X-XYZ # your project_ID
#SBATCH -J job-serial # name of the job
#SBATCH -n *FIXME* # nr. tasks
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
ml purge > /dev/null 2>&1
ml GCC/12.2.0 OpenMPI/4.1.4 R/4.2.2
Rscript --no-save --no-restore integration2d.R
#!/bin/bash
#SBATCH -A lu202X-XX-XX # your project_ID
#SBATCH -J job-serial # name of the job
#SBATCH -n *FIXME* # nr. tasks
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
#SBATCH --reservation=RPJM-course*FIXME* # reservation (optional)
ml purge > /dev/null 2>&1
ml GCC/11.3.0 OpenMPI/4.1.4 R/4.2.1
Rscript --no-save --no-restore integration2d.R
Try different number of cores for this batch script (FIXME string) using the sequence: 1,2,4,8,12, and 14. Note: this number should match the number of processes (also a FIXME string) in the R script. Collect the timings that are printed out in the job.*.out. According to these execution times what would be the number of cores that gives the optimal (fastest) simulation?
Challenge: Increase the grid size (n) to 10000 and submit the batch job with 4 workers (in the
R script) and request 5 cores in the batch script. Monitor the usage of resources
with tools available at your center, for instance top (UPPMAX) or
job-usage (HPC2N).
Here is a parallel code using the parfor tool from Matlab (call it
integration2d.m).
integrationd.m
% Number of workers/processes
num_workers = *FIXME*;
% Use parallel pool with 'parfor'
parpool('kebnekaise',num_workers); % Start parallel pool with num_workers workers
% Grid size
n = 6720;
% bin size
h = pi / n;
tic; % Start timer
% Shared variable to collect partial sums
partial_integrals = 0.0;
% In Matlab one can use parfor to parallelize loops
parfor i = 1:n
partial_integrals = partial_integrals + integration2d_partial(n,i);
end
% Compute the integrals by multilpying by the bin size
integral = partial_integrals * h^2;
elapsedTime = toc; % Stop timer
fprintf("Integral value is %e\n", integral);
fprintf("Error is %e\n", abs(integral - 0.0));
fprintf("Time spent: %.2f sec\n", elapsedTime);
% Clean up the parallel pool
delete(gcp('nocreate'));
% Function for the 2D integration only computes a single bin
function mysum = integration2d_partial(n,i)
% bin size
h = pi / n;
% Partial summation
mysum = 0.0;
% A single bin is computed
x = h * (i - 0.5);
% Regular integration in the Y axis
for j = 1:n
y = h * (j - 0.5);
mysum = mysum + sin(x + y);
end
end
You can run directly this script from the Matlab GUI. Try different number of cores for this batch script (FIXME string) using the sequence: 1,2,4,8,12, and 14. Collect the timings that are printed out in the Matlab command window. According to these execution times what would be the number of cores that gives the optimal (fastest) simulation?
Challenge: Increase the grid size (n) to 100000 and submit the batch job with 4 workers.
Monitor the usage of resources with tools available at your center, for instance top (UPPMAX),
job-usage (HPC2N), or if you’re working in the GUI (e.g. on LUNARC), you can click Parallel
and then Monitor Jobs. For job-usage, you can see the job ID if you type squeue --me on a terminal on Kebnekaise.
Parallelizing a for loop workflow (Advanced)
Create a Data Frame containing two features, one called ID which has integer values from 1 to 10000, and the other called Value that contains 10000 integers starting from 3 and goes in steps of 2 (3, 5, 7, …). The following codes contain parallelized workflows whose goal is to compute the average of the whole feature Value using some number of workers. Substitute the FIXME strings in the following codes to perform the tasks given in the comments.
The main idea for all languages is to divide the workload across all workers. You can run the codes as suggested for each language.
Pandas is available in the following combo ml GCC/12.3.0 SciPy-bundle/2023.07 (HPC2N) and
ml python/3.11.8 (UPPMAX). Call the script script-df.py.
import pandas as pd
import multiprocessing
# Create a DataFrame with two sets of values ID and Value
data_df = pd.DataFrame({
'ID': range(1, 10001),
'Value': range(3, 20002, 2) # Generate 10000 odd numbers starting from 3
})
# Define a function to calculate the sum of a vector
def calculate_sum(values):
total_sum = *FIXME*(values)
return *FIXME*
# Split the 'Value' column into chunks of size 1000
chunk_size = *FIXME*
value_chunks = [data_df['Value'][*FIXME*:*FIXME*] for i in range(0, len(data_df['*FIXME*']), *FIXME*)]
# Create a Pool of 4 worker processes, this is required by multiprocessing
pool = multiprocessing.Pool(processes=*FIXME*)
# Map the calculate_sum function to each chunk of data in parallel
results = pool.map(*FIXME: function*, *FIXME: chunk size*)
# Close the pool to free up resources, if the pool won't be used further
pool.close()
# Combine the partial results to get the total sum
total_sum = sum(results)
# Compute the mean by dividing the total sum by the total length of the column 'Value'
mean_value = *FIXME* / len(data_df['*FIXME*'])
# Print the mean value
print(mean_value)
Run the code with the batch script:
#!/bin/bash -l
#SBATCH -A naiss2024-22-107 # your project_ID
#SBATCH -J job-parallel # name of the job
#SBATCH -n 4 # nr. tasks/coresw
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
# Load any modules you need, here for Python 3.11.8 and compatible SciPy-bundle
module load python/3.11.8
python script-df.py
#!/bin/bash
#SBATCH -A hpc2n202x-XXX # your project_ID
#SBATCH -J job-parallel # name of the job
#SBATCH -n 4 # nr. tasks
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
# Load any modules you need, here for Python 3.11.3 and compatible SciPy-bundle
module load GCC/12.3.0 Python/3.11.3 SciPy-bundle/2023.07
python script-df.py
#!/bin/bash
#SBATCH -A lu202X-XX-XX # your project_ID
#SBATCH -J job-parallel # name of the job
#SBATCH -n 4 # nr. tasks
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
#SBATCH --reservation=RPJM-course*FIXME* # reservation (optional)
# Purge and load any modules you need, here for Python & SciPy-bundle
ml purge
ml GCCcore/12.3.0 Python/3.11.3 SciPy-bundle/2023.07
python dscript-df.py
First, be sure you have
DataFramesinstalled as JuliaPackage.If not, follow the steps below. You can install it in your ordinaty user space (not an environment)
Open a Julia session
julia> using DataFrames
Let it be installed when asking
When done and working, exit().
Here is an exercise to fix some code snippets. Call the script
script-df.jl.Watch out for
*FIXME*and replace with suitable functionsThe functions
nthreads()(number of available threads), andthreadid()(the thread identification number) will be useful in this task.
using DataFrames
using Base.Threads
# Create a data frame with two sets of values ID and Value
data_df = DataFrame(ID = 1:10000, Value = range(3, step=2, length=10000))
# Define a function to compute the sum in parallel
function parallel_sum(data)
# Initialize an array to store thread-local sums
local_sums = zeros(eltype(data), *FIXME*)
# Iterate through each value in the 'Value' column in parallel
@threads for i =1:length(data)
# Add the value to the thread-local sum
local_sums[*FIXME*] += data[i]
end
# Combine the local sums to obtain the total sum
total_sum_parallel = sum(local_sums)
return total_sum_parallel
end
# Compute the sum in parallel
total_sum_parallel = parallel_sum(data_df.Value)
# Compute the mean
mean_value_parallel = *FIXME* / length(data_df.Value)
# Print the mean value
println(mean_value_parallel)
Run this job with the following batch script, defining that we want to use 4 threads:
#!/bin/bash -l
#SBATCH -A naiss2024-22-107 # your project_ID
#SBATCH -J job-parallel # name of the job
#SBATCH -n 4 # nr. tasks/coresw
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
ml julia/1.8.5
julia --threads 4 script-df.jl # X number of threads
#!/bin/bash
#SBATCH -A hpc2n2023-110 # your project_ID
#SBATCH -J job-parallel # name of the job
#SBATCH -n 4 # nr. tasks
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
ml purge > /dev/null 2>&1
ml Julia/1.8.5-linux-x86_64
julia --threads 4 script-df.jl # X number of threads
#!/bin/bash
#SBATCH -A lu202X-XX-XX # your project_ID
#SBATCH -J job-parallel # name of the job
#SBATCH -n 4 # nr. tasks
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
#SBATCH --reservation=RPJM-course*FIXME* # reservation (optional)
ml purge
ml Julia/1.9.3-linux-x86_64
julia --threads 4 script-df.jl # X number of threads
Call the script
script-df.R.
library(doParallel)
library(foreach)
# Create a data frame with two sets called ID and Value
data_df <- data.frame(
ID <- seq(1,10000), Value <- seq(from=3,by=2,length.out=10000)
)
# Create 4 subsets
num_subsets <- *FIXME*
# Create a cluster with 4 workers
cl <- makeCluster(*FIXME*)
# Register the cluster for parallel processing
registerDoParallel(cl)
# Function to process a subset of the whole data
process_subset <- function(subset) {
# Perform some computation on the subset
subset_sum <- sum(*FIXME*)
return(data.frame(SubsetSum = subset_sum))
}
# Use foreach with dopar to process subsets in parallel
result <- foreach(i = 1:*FIXME*, .combine = rbind) %dopar% {
# Determine the indices for the subset
subset_indices <- seq(from = *FIXME*,
to = *FIXME*)
# Create the subset
subset_data <- data_df[*FIXME*, , drop = FALSE]
# Process the subset
subset_result <- process_subset(*FIXME*)
return(subset_result)
}
# Stop the cluster when done
stopCluster(cl)
# Print the results
print(sum(*FIXME*)/*FIXME*)
Run the code with the following batch script:
#!/bin/bash -l
#SBATCH -A naiss2024-22-107 # your project_ID
#SBATCH -J job-parallel # name of the job
#SBATCH -n 4 # nr. tasks/coresw
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
ml R_packages/4.1.1
Rscript --no-save --no-restore script-df.R
#!/bin/bash
#SBATCH -A hpc2n202X-XYZ # your project_ID
#SBATCH -J job-parallel # name of the job
#SBATCH -n 4 # nr. tasks
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
ml purge > /dev/null 2>&1
ml GCC/12.2.0 OpenMPI/4.1.4 R/4.2.2
Rscript --no-save --no-restore script-df.R
#!/bin/bash
#SBATCH -A lu202X-XX-XX # your project_ID
#SBATCH -J job-parallel # name of the job
#SBATCH -n 4 # nr. tasks
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
#SBATCH --reservation=RPJM-course*FIXME* # reservation (optional)
ml purge > /dev/null 2>&1
ml GCC/11.3.0 OpenMPI/4.1.4 R/4.2.1
Rscript --no-save --no-restore script-df.R
% Create a table with two columns: ID and Value ID = (1:10000)'; % Column for IDs Value = (3:2:20001)'; % Column for values data_tbl = table(*FIXME*, *FIXME*); % Create a table with the previous two features % Matlab uses the so called parpool to create some workers parpool('kebnekaise', *FIXME*); p = gcp; % Measure time tic; % Compute the sum in parallel for the Value feature total_sum_parallel = parallel_sum(data_tbl.*FIXME*); % Compute the mean mean_value_parallel = total_sum_parallel / length(data_tbl.*FIXME*); % Stop measuring time t_parallel = toc; fprintf('Time taken for parallel version: %.2f seconds\n', t_parallel); % Display the mean value disp(mean_value_parallel); % Delete the pool delete(gcp); % Function to compute the sum in parallel function total_sum_parallel = parallel_sum(values) n = length(*FIXME*); local_sums = 0.0; parfor i = 1:*FIXME* % run the loop over the number of elements local_sums = local_sums + *FIXME*(i); % add the values to the partial sum end % Set the total sum total_sum_parallel = local_sums; end
You can run this code directly from the Matlab GUI.
Solution
import pandas as pd
import multiprocessing
# Create a DataFrame with two sets of values ID and Value
data_df = pd.DataFrame({
'ID': range(1, 10001),
'Value': range(3, 20002, 2) # Generate 10000 odd numbers starting from 3
})
# Define a function to calculate the sum of a vector
def calculate_sum(values):
total_sum = sum(values)
return total_sum
# Split the 'Value' column into chunks
chunk_size = 1000
value_chunks = [data_df['Value'][i:i+chunk_size] for i in range(0, len(data_df['Value']), chunk_size)]
# Create a Pool of 4 worker processes, this is required by multiprocessing
pool = multiprocessing.Pool(processes=4)
# Map the calculate_sum function to each chunk of data in parallel
results = pool.map(calculate_sum, value_chunks)
# Close the pool to free up resources, if the pool won't be used further
pool.close()
# Combine the partial results to get the total sum
total_sum = sum(results)
# Compute the mean by dividing the total sum by the total length of the column 'Value'
mean_value = total_sum / len(data_df['Value'])
# Print the mean value
print(mean_value)
using DataFrames
using Base.Threads
# Create a data frame with two sets of values ID and Value
data_df = DataFrame(ID = 1:10000, Value = range(3, step=2, length=10000))
# Define a function to compute the sum in parallel
function parallel_sum(data)
# Initialize an array to store thread-local sums
local_sums = zeros(eltype(data), nthreads())
# Iterate through each value in the 'Value' column in parallel
@threads for i =1:length(data)
# Add the value to the thread-local sum
local_sums[threadid()] += data[i]
end
# Combine the local sums to obtain the total sum
total_sum_parallel = sum(local_sums)
return total_sum_parallel
end
# Compute the sum in parallel
total_sum_parallel = parallel_sum(data_df.Value)
# Compute the mean
mean_value_parallel = total_sum_parallel / length(data_df.Value)
# Print the mean value
println(mean_value_parallel)
library(doParallel)
library(foreach)
# Create a data frame with two sets called ID and Value
data_df <- data.frame(
ID <- seq(1,10000), Value <- seq(from=3,by=2,length.out=10000)
)
# Create 4 subsets
num_subsets <- 4
# Create a cluster with 4 workers
cl <- makeCluster(4)
# Register the cluster for parallel processing
registerDoParallel(cl)
# Function to process a subset of the whole data
process_subset <- function(subset) {
# Perform some computation on the subset
subset_sum <- sum(subset$Value)
return(data.frame(SubsetSum = subset_sum))
}
# Use foreach with dopar to process subsets in parallel
result <- foreach(i = 1:num_subsets, .combine = rbind) %dopar% {
# Determine the indices for the subset
subset_indices <- seq(from = 1 + (i - 1) * nrow(data_df) / num_subsets,
to = i * nrow(data_df) / num_subsets)
# Create the subset
subset_data <- data_df[subset_indices, , drop = FALSE]
# Process the subset
subset_result <- process_subset(subset_data)
return(subset_result)
}
# Stop the cluster when done
stopCluster(cl)
# Print the results
print(sum(result)/10000)
% Create a table with two columns: ID and Value
ID = (1:10000)'; % Column for IDs
Value = (3:2:20001)'; % Column for values
data_tbl = table(ID, Value);
% Matlab uses the so called parpool to create some workers
parpool('kebnekaise', 4);
p = gcp;
% Measure time
tic;
% Compute the sum in parallel
total_sum_parallel = parallel_sum(data_tbl.Value);
% Compute the mean
mean_value_parallel = total_sum_parallel / length(data_tbl.Value);
% Stop measuring time
t_parallel = toc;
fprintf('Time taken for parallel version: %.2f seconds\n', t_parallel);
% Display the mean value
disp(mean_value_parallel);
% Delete the pool
delete(gcp);
% Function to compute the sum in parallel
function total_sum_parallel = parallel_sum(values)
n = length(values);
local_sums = 0.0;
parfor i = 1:n
local_sums = local_sums + values(i);
end
% Set the total sum
total_sum_parallel = local_sums;
end
More info
Introduction to Dask by Aalto Scientific Computing and CodeRefinery
The book High Performance Python is a good resource for ways of speeding up Python code.