Introduction to running R, Python, Julia, and Matlab in HPC
Content
This course aims to give a brief, but comprehensive introduction to using Python, Julia, R, and Matlab in an HPC environment.
- You will learn how to
start the interpreters of Python, Julia, R, and Matlab by the HPC module system
find site installed packages/libraries
install packages/libraries yourself
use virtual environments
use the computation nodes
write and submit batch scripts
work interactivly
Learn about the batch system
Learn about GPUs and Python
access parallel tools and run parallel codes
This course will consist of lectures interspersed with hands-on sessions where you get to try out what you have just learned.
We aim to give this course in spring and fall every year.
Course approach to deal with multiple HPC centers
The course is a cooperation between UPPMAX (Rackham, Snowy, Bianca), HPC2N (Kebnekaise), and LUNARC (Cosmos) and will focus on the compute systems at all three centres.
Although there are differences we will only have few seperate sessions.
Most participants will use UPPMAX’s systems for the course, as Kebnekaise and Cosmos are only for local users. Kebnekaise: UmU, IRF, MIUN, SLU, LTU. Cosmos: LU.
The general information given in the course will be true for all/most HPC centres in Sweden.
The examples will often have specific information, like module names and versions, which may vary. What you learn here should help you to make any changes needed for the other centres.
When present, links to the Python/Julia/R/Matlab documentation at other NAISS centres are given in the corresponding session.
Schedule Fall 2024
Day |
Language |
|---|---|
Tuesday 22 October |
Python |
Wednesday 23 October |
Julia |
Thursday 24 October |
R |
Friday 25 October |
Matlab |
Some practicals
- Code of Conduct
Be nice to each other!
- Zoom
You should have gotten an email with the links
Zoom policy:
Zoom chat (maintained by co-teachers):
technical issues of zoom
technical issues of your settings
direct communication
each teacher may have somewhat different approach
collaboration document (see below):
“explain again”
elaborating the course content
solutions for your own work
- Recording policy:
The lectures and demos will be recorded.
The questions asked per microphone during these sessions will be recorded
If you don’t want your voice to appear use:
use the collaboration document (see below)
The Zoom main room is used for most lectures
Some sessions use breakout rooms for exercises, some of which use a silent room
Collaboration document
- Q/A collaboration document
Use this page for the workshop with your questions
It helps us identify content that is missing in the course material
We answer those questions as soon as possible
Warning
Please be sure that you have gone through the `pre-requirements <https://uppmax.github.io/R-python-julia-matlab-HPC/prereqs.html>`_
It mentions the familiarity with the LINUX command line.
- The applications to connect to the clusters
terminals
remote graphical desktop ThinLinc
Material for improving your programming skills
This course does not aim to improve your coding skills.
Rather you will learn to understand the ecosystems and navigations for the the different languages on a HPC cluster.
Briefly about the cluster hardware and system at UPPMAX and HPC2N
What is a cluster?
Login nodes and calculations/computation nodes
A network of computers, each computer working as a node.
Each node contains several processor cores and RAM and a local disk called scratch.
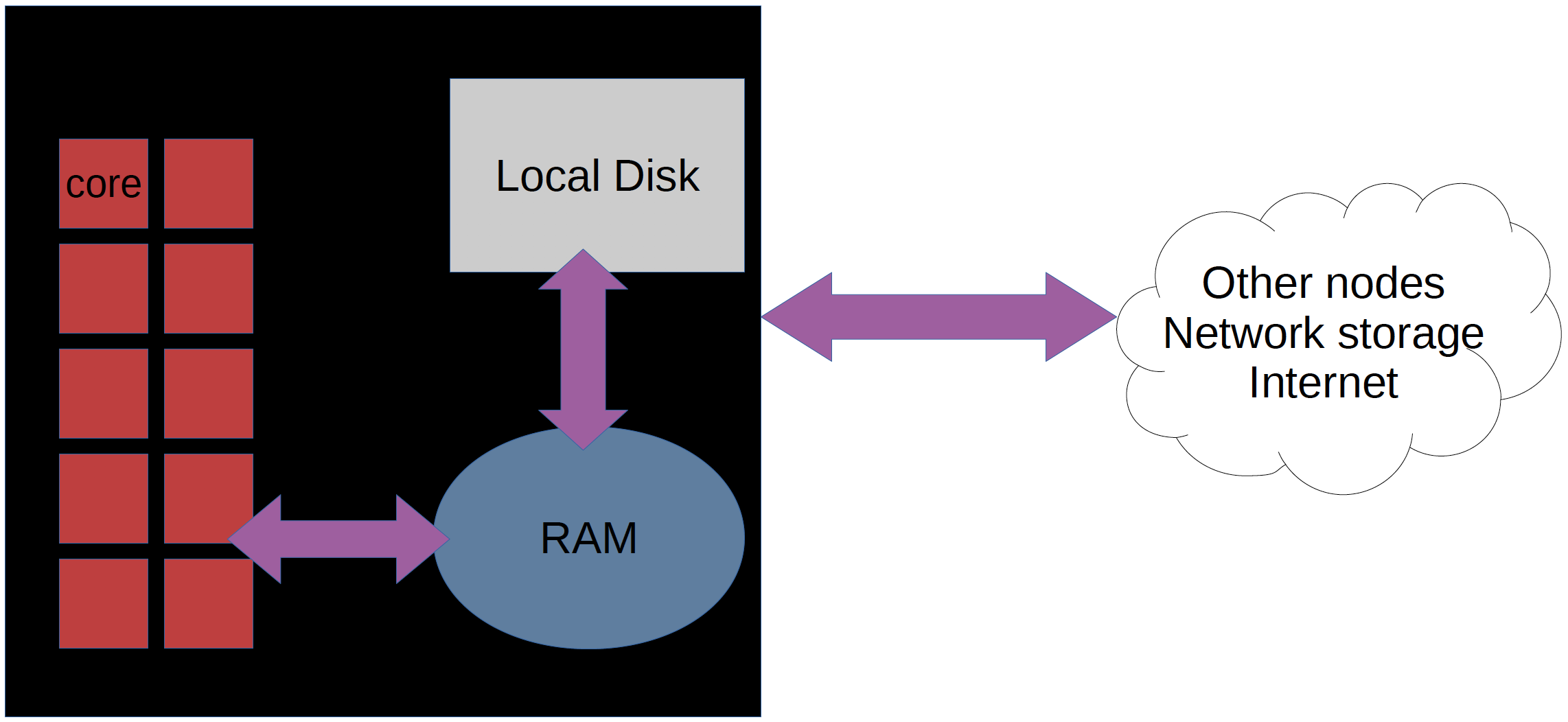
The user logs in to login nodes via Internet through ssh or Thinlinc.
Here the file management and lighter data analysis can be performed.
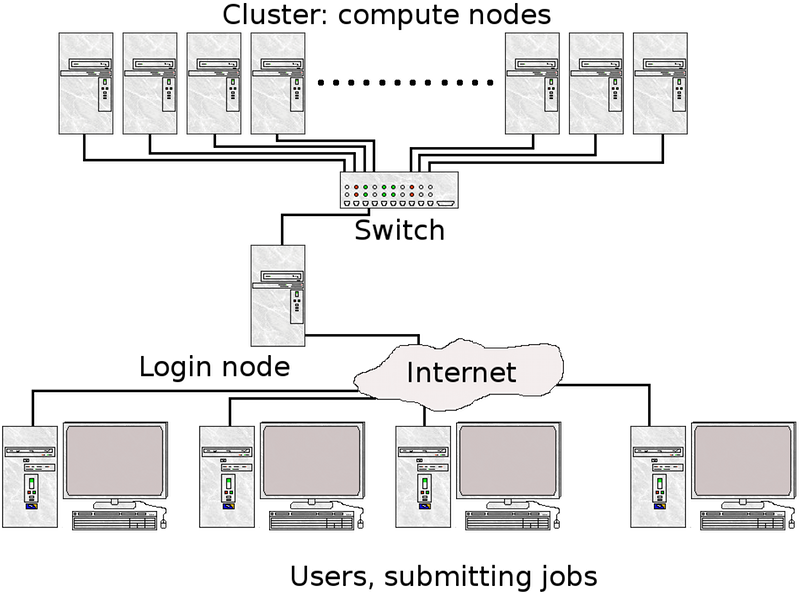
The calculation nodes have to be used for intense computing.
The HPC centers UPPMAX, HPC2N, and LUNARC
Three HPC centers
There are many similarities:
Login vs. calculation/compute nodes
Environmental module system with software hidden until loaded with
module loadSlurm batch job and scheduling system
pip installprocedure
… and small differences:
commands to load Python, Python packages, R, Julia, and MATLAB
slightly different flags to Slurm
… and some bigger differences:
UPPMAX has three different clusters:
Rackham for general purpose computing on CPUs only
Snowy available for local projects and suits long jobs (< 1 month) and has GPUs
Bianca for sensitive data and has GPUs
HPC2N has Kebnekaise with GPUs.
LUNARC has Cosmos, with 9 GPUs (4 are partitions of Intel nodes).
LUNARC has Desktop On Demand, allowing some applications to run on the back-end without the use of a terminal or batch script.
Conda is recommended only for UPPMAX and LUNARC users, and only using the conda-forge repository.
Prepare your environment now!
Log in and create a user folder (if not done already)
Please log in to Rackham, Kebnekaise, Cosmos or other cluster that you are using.
Use Thinlinc or terminal?
It is up to you!
Graphics come easier with Thinlinc
For this course, when having many windows open, it may sometimes be better to run in terminal, for screen space issues.
Log in to Rackham!
Terminal:
ssh -X <user>@rackham.uppmax.uu.seThinLinc app:
server:
rackham-gui.uppmax.uu.seusername:
<user>ThinLinc in web browser:
https://rackham-gui.uppmax.uu.se
If not already: create a working directory where you can code along.
We recommend creating it under the course project storage directory
Example. If your username is “mrspock” and you are at UPPMAX, then we recommend you create this folder:
$ mkdir /proj/r-py-jl-m-rackham/mrspock/
Kebnekaise through terminal:
<user>@kebnekaise.hpc2n.umu.seKebnekaise through ThinLinc app
server:
kebnekaise-tl.hpc2n.umu.seusername:
<user>
Create a working directory where you can code along.
Example. If your username is
bbrydsoeand you are at HPC2N, then we recommend you create this folder:
$ /proj/nobackup/r-py-jl-m/bbrydsoe/
Cosmos through terminal:
<user>@cosmos.lunarc.lu.seCosmos through ThinLinc app, (requires 2FA)
server:
cosmos-dt.lunarc.lu.seusername:
<user>
Create a working directory where you can code along. Users should have plenty of space in their home directories.
Get exercises
There are three main ways to get the exercises. In any case, you should do so from the directory you will be working in, on either Rackham, Kebnekaise, or Cosmos:
Copy them from the computer system you are on (only until 2024-11-01) and unpack them by entering
tar -xzvf exercises.tar.gzRackham:
cp /proj/r-py-jl-m-rackham/exercises.tar.gz.Kebnekaise:
cp /proj/nobackup/r-py-jl-m/exercises.tar.gz.(No local repository for Cosmos)
Clone them with git from the repo (see below about a warning):
git clone https://github.com/UPPMAX/R-python-julia-matlab-HPC.gitCopy the tarball from the web into your working directory with
wget https://github.com/UPPMAX/R-python-julia-matlab-HPC/raw/refs/heads/main/exercises/exercises.tar.gzand unpack them withtar -xzvf exercises.tar.gz
Warning
Do you want the whole repo?
If you are happy with just the exercises, the tarballs of the language specific ones are enough.
By cloning the whole repo, you get all the materials, planning documents, and exercises.
If you think this makes sense type this in the command line in the directory you want it. -
git clone https://github.com/UPPMAX/R-python-julia-matlab-HPC.git- Note however, that if you during exercise work modify files, they will be overwritten if you make
git pull(like if the teacher needs to modify something). Then make a copy somewhere else with your answers!
- Note however, that if you during exercise work modify files, they will be overwritten if you make
Summary of Project ID and directory name
Main project on UPPMAX:
Project ID:
naiss2024-22-1202Directory name on rackham:
/proj/r-py-jl-m-rackhamPlease create a suitably named subdirectory below
/proj/r-py-jl-m-rackham, for your own exercises.
Local project on HPC2N:
Project ID:
hpc2n2024-114Directory name on Kebnekaise:
/proj/nobackup/r-py-jl-mPlease create a suitably named subdirectory below
/proj/nobackup/r-py-jl-m, for your own exercises.
Where to work on LUNARC
Project ID:
lu2024-7-80(for use in slurm scripts)Home directories have much larger quotas at LUNARC than at UPPMAX or HPC2N. Create a suitable sub-directory in your home directory or a personal project folder.
Content of the course
Pre-requirements:
COMMON:
Python Lessons:
- Introduction Python
- Load and run python
- Python packages
- Isolated environments
- Running Python in batch mode
- Using GPUs with Python
- Interactive work on the compute nodes
- Introduction
- The different way HPC2N, UPPMAX, and LUNARC provide for an interactive session
- Start an interactive session
- Check to be in an interactive session
- Check to have booked the expected amount of cores
- Running a Python script in an interactive session
- End the interactive session
- Exercises
- Exercise 0: be able to use the Python scripts
- Exercise 1
- Exercise 2
- Conclusion
- Links
- Jupyter on compute nodes
- Parallel and multithreaded functions
- Conda at UPPMAX
- Summary
- Evaluation
Julia Lessons:
R Lessons:
Matlab Lessons: