Running R in batch mode
Questions
What is a batch job?
How to write a batch script and submit a batch job?
Objectives
Short introduction to SLURM scheduler
Show structure of a batch script
Examples to try
Compute allocations in this workshop
Rackham:
naiss2024-22-107Kebnekaise:
hpc2n2024-025Cosmos:
lu2024-7-80
Storage space for this workshop
Rackham:
/proj/r-py-jl-m-rackhamKebnekaise:
/proj/nobackup/r-py-jl-m
Overview of the UPPMAX systems
Overview of the HPC2N system
Overview of the LUNARC system
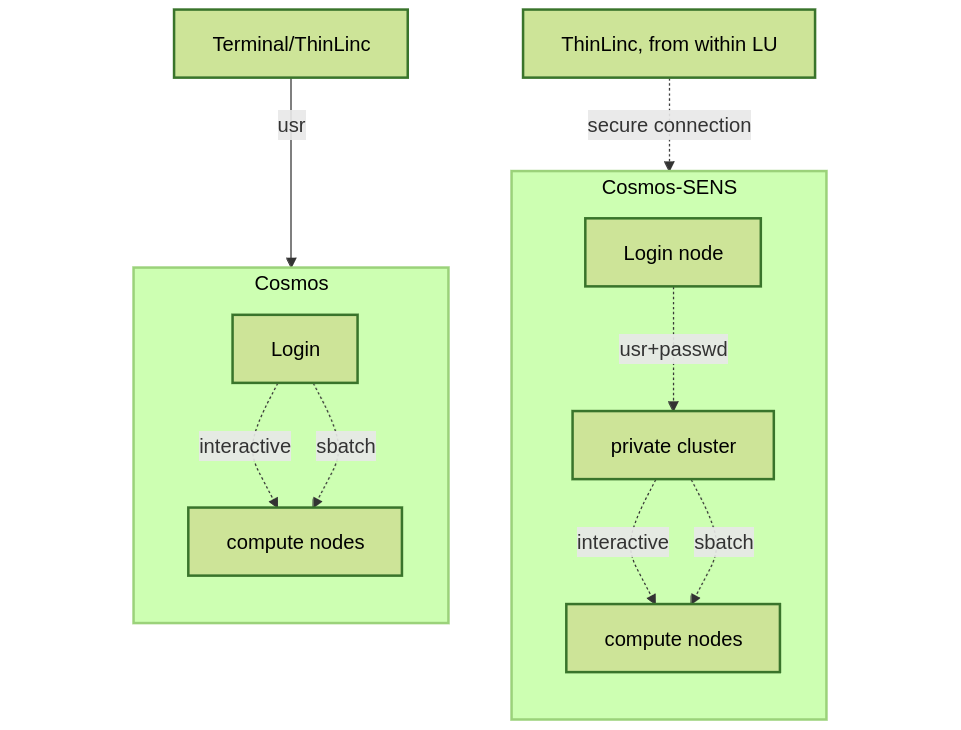
Any longer, resource-intensive, or parallel jobs must be run through a batch script.
The batch system used at UPPMAX, HPC2N, and LUNARC (and most other HPC centres in Sweden) is called Slurm.
Slurm is an Open Source job scheduler, which provides three key functions
Keeps track of available system resources
Enforces local system resource usage and job scheduling policies
Manages a job queue, distributing work across resources according to policies
In order to run a batch job, you need to create and submit a SLURM submit file (also called a batch submit file, a batch script, or a job script).
Guides and documentation at: https://docs.hpc2n.umu.se/documentation/batchsystem/intro/ and https://docs.uppmax.uu.se/cluster_guides/slurm/ and https://lunarc-documentation.readthedocs.io/en/latest/manual/manual_intro/
Workflow
Write a batch script
Inside the batch script you need to load the modules you need (R and any prerequisites)
If you are using any own-installed packages, make sure R_LIBS_USER is set (export R_LIBS_USER=/path/to/my/R-packages)
Ask for resources depending on if it is a parallel job or a serial job, if you need GPUs or not, etc.
Give the command(s) to your R script
Submit batch script with
sbatch <my-batch-script-for-R.sh>
Common file extensions for batch scripts are .sh or .batch, but they are not necessary. You can choose any name that makes sense to you.
Useful commands to the batch system
Submit job:
sbatch <jobscript.sh>Get list of your jobs:
squeue -u <username>Check on a specific job:
scontrol show job <job-id>Delete a specific job:
scancel <job-id>Useful info about a job:
sacct -l -j <job-id> | less -SUrl to a page with info about the job (Kebnekaise only):
job-usage <job-id>
Keypoints
The Slurm scheduler handles allocations to the calculation nodes
Interactive sessions was presented in the previous presentation
Batch jobs runs without interaction with the user
- A batch script consists of a part with Slurm parameters describing the allocation and a second part describing the actual work within the job, for instance one or several R scripts.
Remember to include possible input arguments to the R script in the batch script.
Example R batch scripts
Serial code
Type-Along
Short serial batch example for running the code hello.R
Short serial example script for Rackham. Loading R/4.1.1
#!/bin/bash
#SBATCH -A naiss2024-22-1202 # Course project id. Change to your own project ID after the course
#SBATCH --time=00:10:00 # Asking for 10 minutes
#SBATCH -n 1 # Asking for 1 core
# Load any modules you need, here R/4.1.1
module load R/4.1.1
# Run your R script (here 'hello.R')
R --no-save --quiet < hello.R
Short serial example for running on Kebnekaise. Loading R/4.1.2 and prerequisites
#!/bin/bash
#SBATCH -A hpc2n2024-114 # Change to your own project ID
#SBATCH --time=00:10:00 # Asking for 10 minutes
#SBATCH -n 1 # Asking for 1 core
# Load any modules you need, here R/4.1.2 and prerequisites
module load GCC/11.2.0 OpenMPI/4.1.1 R/4.1.2
# Run your R script (here 'hello.R')
R --no-save --quiet < hello.R
Short serial example for running on Cosmos. Loading R/4.2.1 and prerequisites
#!/bin/bash
#SBATCH -A lu2024-7-80 # Change to your own project ID
#SBATCH --time=00:10:00 # Asking for 10 minutes
#SBATCH -n 1 # Asking for 1 core
# Load any modules you need, here R/4.1.2 and prerequisites
module load GCC/11.3.0 OpenMPI/4.1.4 R/4.2.1
# Run your R script (here 'hello.R')
R --no-save --quiet < hello.R
R example code
message <-"Hello World!"
print(message)
Send the script to the batch:
$ sbatch <batch script>
Parallel code
foreach and doParallel
Type-Along
Short parallel example, using foreach and doParallel
Short parallel example (Since we are using packages “foreach” and “doParallel”, you need to use module R_packages/4.1.1 instead of R/4.1.1.
#!/bin/bash
#SBATCH -A naiss2024-22-1202
#SBATCH -t 00:10:00
#SBATCH -N 1
#SBATCH -c 4
ml purge > /dev/null 2>&1
ml R_packages/4.1.1
# Batch script to submit the R program parallel_foreach.R
R -q --slave -f parallel_foreach.R
Short parallel example (using packages “foreach” and “doParallel” which are included in the R module) for running on Kebnekaise. Loading R/4.0.4 and its prerequisites.
#!/bin/bash
#SBATCH -A hpc2n2024-114 # Change to your own project ID
#SBATCH -t 00:10:00
#SBATCH -N 1
#SBATCH -c 4
ml purge > /dev/null 2>&1
ml GCC/10.2.0 OpenMPI/4.0.5 R/4.0.4
# Batch script to submit the R program parallel_foreach.R
R -q --slave -f parallel_foreach.R
Short parallel example (using packages “foreach” and “doParallel” which are included in the R module) for running on Cosmos. Loading R/4.2.1 and its prerequisites.
#!/bin/bash
# A batch script for running the R program parallel_foreach.R on Kebnekaise
#SBATCH -A lu2024-7-80 # Change to your own project ID
#SBATCH -t 00:10:00
#SBATCH -N 1
#SBATCH -c 4
ml purge > /dev/null 2>&1
ml GCC/11.3.0 OpenMPI/4.1.4 R/4.2.1
# Batch script to submit the R program parallel_foreach.R
R -q --slave -f parallel_foreach.R
This R script uses packages “foreach” and “doParallel”.
library(parallel)
library(foreach)
library(doParallel)
# Function for calculating PI with no values
calcpi <- function(no) {
y <- runif(no)
x <- runif(no)
z <- sqrt(x^2+y^2)
length(which(z<=1))*4/length(z)
}
# Detect the number of cores
no_cores <- detectCores() - 1
# Loop to max number of cores
for (n in 1:no_cores) {
# print how many cores we are using
print(n)
# Set start time
start_time <- Sys.time()
# Create a cluster
nproc <- makeCluster(n)
registerDoParallel(nproc)
# Create a vector 1000 length with 100 randomizations
input <- rep(100, 1000)
# Use foreach on n cores
registerDoParallel(nproc)
res <- foreach(i = input, .combine = '+') %dopar%
calcpi(i)
# Print the mean of the results
print(res/length(input))
# Stop the cluster
stopCluster(nproc)
# print end time
print(Sys.time() - start_time)
}
Send the script to the batch:
$ sbatch <batch script>
Rmpi
Type-Along
Short parallel example using package “Rmpi”
Short parallel example (using package “Rmpi”, so we need to load the module R_packages/4.1.1 instead of R/4.1.1 and we need to load a suitable openmpi module, openmpi/4.0.3)
#!/bin/bash
#SBATCH -A naiss2024-22-1202
#Asking for 10 min.
#SBATCH -t 00:10:00
#SBATCH -n 8
export OMPI_MCA_mpi_warn_on_fork=0
export OMPI_MCA_btl_openib_allow_ib=1
ml purge > /dev/null 2>&1
ml R_packages/4.1.1
ml openmpi/4.0.3
mpirun -np 1 R CMD BATCH --no-save --no-restore Rmpi.R output.out
Short parallel example (using packages “Rmpi”). Loading R/4.0.4 and its prerequisites.
#!/bin/bash
#SBATCH -A hpc2n2024-114 # Change to your own project ID
#Asking for 10 min.
#SBATCH -t 00:10:00
#SBATCH -n 8
export OMPI_MCA_mpi_warn_on_fork=0
ml purge > /dev/null 2>&1
ml GCC/10.2.0 OpenMPI/4.0.5
ml R/4.1.1
mpirun -np 1 R CMD BATCH --no-save --no-restore Rmpi.R output.out
Short parallel example (using packages “Rmpi”). Loading R/4.0.4 and its prerequisites.
#!/bin/bash
#SBATCH -A lu2024-7-80 # Change to your own project ID
# Asking for 10 min.
#SBATCH -t 00:10:00
#SBATCH -n 8
export OMPI_MCA_mpi_warn_on_fork=0
ml purge > /dev/null 2>&1
ml GCC/11.3.0 OpenMPI/4.1.4
ml R/4.2.1
mpirun -np 1 R CMD BATCH --no-save --no-restore Rmpi.R output.out
This R script uses package “Rmpi”.
# Load the R MPI package if it is not already loaded.
if (!is.loaded("mpi_initialize")) {
library("Rmpi")
}
print(mpi.universe.size())
ns <- mpi.universe.size() - 1
mpi.spawn.Rslaves(nslaves=ns)
#
# In case R exits unexpectedly, have it automatically clean up
# resources taken up by Rmpi (slaves, memory, etc...)
.Last <- function(){
if (is.loaded("mpi_initialize")){
if (mpi.comm.size(1) > 0){
print("Please use mpi.close.Rslaves() to close slaves.")
mpi.close.Rslaves()
}
print("Please use mpi.quit() to quit R")
.Call("mpi_finalize")
}
}
# Tell all slaves to return a message identifying themselves
mpi.remote.exec(paste("I am",mpi.comm.rank(),"of",mpi.comm.size(),system("hostname",intern=T)))
# Test computations
x <- 5
x <- mpi.remote.exec(rnorm, x)
length(x)
x
# Tell all slaves to close down, and exit the program
mpi.close.Rslaves()
mpi.quit()
Send the script to the batch system:
$ sbatch <batch script>
Using GPUs in a batch job
There are generally either not GPUs on the login nodes or they cannot be accessed for computations. To use them you need to either launch an interactive job or submit a batch job.
UPPMAX only
Rackham’s compute nodes do not have GPUs. You need to use Snowy for that.
You need to use this batch command (for x being the number of cards, 1 or 2):
#SBATCH -M snowy
#SBATCH --gres=gpu:x
HPC2N
Kebnekaise’s GPU nodes are considered a separate resource, and the regular compute nodes do not have GPUs.
Kebnekaise has a great many different types of GPUs:
V100 (2 cards/node)
A40 (8 cards/node)
A6000 (2 cards/node)
L40s (2 or 6 cards/node)
A100 (2 cards/node)
H100 (4 cards/node)
MI100 (2 cards/node)
To access them, you need to use this to the batch system:
#SBATCH --gpus=x
where x is the number of GPU cards you want. Above are given how many are on each type, so you can ask for up to that number.
In addition, you need to add this to the batch system:
#SBATCH -C <type>
where type is
v100
a40
a6000
l40s
a100
h100
mi100
For more information, see HPC2N’s guide to the different parts of the batch system: https://docs.hpc2n.umu.se/documentation/batchsystem/resources/
LUNARC
LUNARC has Nvidia A100 GPUs and Nvidia A40 GPUs, but the latter ones are reserved for interactive graphics work on the on-demand system, and Slurm jobs should not be submitted to them.
Thus in order to use the A100 GPUs on Cosmos, add this to your batch script:
A100 GPUs on AMD nodes:
#SBATCH -p gpua100
#SBATCH --gres=gpu:1
These nodes are configured as exclusive access and will not be shared between users. User projects will be charged for the entire node (48 cores). A job on a node will also have access to all memory on the node.
A100 GPUs on Intel nodes:
#SBATCH -p gpua100i #SBATCH --gres=gpu:<number>
where <number> is 1 or 2 (Two of the nodes have 1 GPU and two have 2 GPUs).
Example batch script
#!/bin/bash
#SBATCH -A naiss2024-22-1202
#Asking for runtime: hours, minutes, seconds. At most 1 week
#SBATCH -t HHH:MM:SS
#SBATCH --exclusive
#SBATCH -p node
#SBATCH -N 1
#SBATCH -M snowy
#SBATCH --gpus=1
#SBATCH --gpus-per-node=1
#Writing output and error files
#SBATCH --output=output%J.out
#SBATCH --error=error%J.error
ml purge > /dev/null 2>&1
ml R/4.1.1 R_packages/4.1.1
R --no-save --no-restore -f MY-R-GPU-SCRIPT.R
#!/bin/bash
#SBATCH -A hpc2n2024-114 # Change to your own project ID
#Asking for runtime: hours, minutes, seconds. At most 1 week
#SBATCH -t HHH:MM:SS
#Ask for GPU resources. You pick type as one of the ones shown above
#x is how many cards you want, at most as many as shown above
#SBATCH --gpus:x
#SBATCH -C type
#Writing output and error files
#SBATCH --output=output%J.out
#SBATCH --error=error%J.error
ml purge > /dev/null 2>&1
#R version 4.0.4 is the only one compiled for CUDA
ml GCC/10.2.0 CUDA/11.1.1 OpenMPI/4.0.5
ml R/4.0.4
R --no-save --no-restore -f MY-R-GPU-SCRIPT.R
#!/bin/bash
# Remember to change this to your own project ID after the course!
#SBATCH -A lu2024-7-80
# Asking for runtime: hours, minutes, seconds. At most 1 week
#SBATCH --time=HHH:MM:SS
# Ask for GPU resources - x is how many cards, 1 or 2
#SBATCH -p gpua100
#SBATCH --gres=gpu:x
# Remove any loaded modules and load the ones we need
module purge > /dev/null 2>&1
module load GCC/11.3.0 OpenMPI/4.1.4 R/4.2.1 CUDA/12.1.1
R --no-save --no-restore -f MY-R-GPU-SCRIPT.R
Exercises
Serial batch script for R
Run the serial batch script from further up on the page, but for the add2.R code. Remember the arguments.
Solution for UPPMAX
Serial script on Rackham
#!/bin/bash #SBATCH -A naiss2024-22-1202 # Change to your own after the course #SBATCH --time=00:10:00 # Asking for 10 minutes #SBATCH -n 1 # Asking for 1 core # Load any modules you need, here for R/4.1.1 module load R/4.1.1 # Run your R script Rscript add2.R 2 3
Solution for HPC2N
Serial script on Kebnekaise
#!/bin/bash #SBATCH -A hpc2n2024-114 # Change to your own project ID #SBATCH --time=00:10:00 # Asking for 10 minutes #SBATCH -n 1 # Asking for 1 core # Load any modules you need, here for R/4.1.2 module load GCC/11.2.0 OpenMPI/4.1.1 R/4.1.2 # Run your R script Rscript add2.R 2 3
Solution for LUNARC
Serial script on R
#!/bin/bash #SBATCH -A lu2024-7-80 # Change to your own project ID #SBATCH --time=00:10:00 # Asking for 10 minutes #SBATCH -n 1 # Asking for 1 core # Load any modules you need, here for R/4.2.1 module load GCC/11.3.0 OpenMPI/4.1.4 R/4.2.1 # Run your R script Rscript add2.R 2 3
Parallel job run
Try making a batch script for running the parallel example with “foreach” from further up on the page.