Parallel computing with Python
Questions
What is parallel computing?
What are the different parallelization mechanisms for Python?
How to implement parallel algorithms in Python code?
How to deploy threads and workers at our HPC centers?
Objectives
Learn general concepts for parallel computing
Gain knowledge on the tools for parallel programming
Familiarize with the tools to monitor the usage of resources
Get today’s tarball!
Prerequisites
Warning
Instructions are updated mainly for the NAISS cluster Tetralith
Demo
These guidelines are working for Tetralith:
$ ml buildenv-gcccuda/12.2.2-gcc11-hpc1
$ python -m venv /path-to-your-project/vpyenv-python-course
$ source /path-to-your-project/vpyenv-python-course/bin/activate
For the
mpi4pyexample add the following modules:
$ pip install mpi4py
For the
numbaexample install the corresponding module:
$ pip install numba
For the Julia example we will need PyJulia:
$ ml julia/1.10.2-bdist
$ pip install JuliaCall
Start Julia on the command line and add the following package:
pkg> add PythonCall
For the
Heatexamples:
$ pip install torch --index-url https://download.pytorch.org/whl/cu126
$ pip install heat[hdf5,netcdf]
The PyOMP example needs an additional package:
$ pip install pyomp
Quit Python, you should be ready to go!
Warning
These instructions work for the AMD nodes only. For the Intel nodes, one needs a different Python version and tool chains.
If not already done so:
$ ml GCCcore/11.2.0 Python/3.9.6
$ python -m venv vpyenv-python-course
$ source /proj/nobackup/<your-project-storage>/vpyenv-python-course/bin/activate
For the
numbaexample install the corresponding module:
$ pip install numba
For the
mpi4pyexample add the following modules:
$ ml GCC/11.2.0 OpenMPI/4.1.1
$ pip install mpi4py
For the Julia example we will need PyJulia:
$ ml Julia/1.9.3-linux-x86_64
$ pip install julia
Start Python on the command line and type:
>>> import julia
>>> julia.install()
This will install the PyCall connector between Python and Julia.
For the
Heatexamples:
$ ml GCC/11.2.0 OpenMPI/4.1.1
$ ml CUDA/12.0.0
$ pip install torch --index-url https://download.pytorch.org/whl/cu126
$ pip install heat[hdf5,netcdf]
The PyOMP example needs an additional package:
$ pip install pyomp
Quit Python, you should be ready to go!
If not already done so:
$ module load python/3.9.5
$ python -m venv --system-site-packages /proj/naiss202X-XY-XYZ/nobackup/<user>/venv-python-course
Activate it if needed (is the name shown in the prompt)
$ source /proj/naiss202X-XY-XYZ/nobackup/<user>/venv-python-course/bin/activate
For the
numbaexample install the corresponding module:
$ python -m pip install numba
For the
mpi4pyexample add the following modules:
$ ml gcc/9.3.0 openmpi/3.1.5
$ python -m pip install mpi4py
For the Julia example we will need PyJulia:
$ ml julia/1.7.2
$ python -m pip install julia
Start Python on the command line and type:
>>> import julia
>>> julia.install()
Quit Python, you should be ready to go!
These guidelines are working for Cosmos:
$ ml GCC/12.3.0 Python/3.11.3
$ ml OpenMPI/4.1.5
$ python -m venv /path-to-your-project/vpyenv-python-course
$ source /path-to-your-project/vpyenv-python-course/bin/activate
For the
mpi4pyexample add the following modules:
$ pip install mpi4py
For the
numbaexample install the corresponding module:
$ pip install numba
For the Julia example we will need PyJulia:
$ ml Julia/1.10.4-linux-x86_64
$ pip install JuliaCall
Start Julia on the command line and add the following package:
# go to package mode
pkg> add PythonCall
# return to Julian mode
julia>using PythonCall
julia>exit()
If not already done so:
$ ml cray-python
$ python -m venv vpyenv-python-course
$ source /proj/nobackup/<your-project-storage>/vpyenv-python-course/bin/activate
For the
numbaexample install the corresponding module:
$ pip install numba
For the
mpi4pyexample add the following modules:
$ pip install mpi4py
Quit Python, you should be ready to go!
What is parallel programming?
Parallel programming is the science and art of writing code that execute tasks on different computing units (cores) simultaneously. In the past computers were shiped with a single core per Central Processing Unit (CPU) and therefore only a single computation at the time (serial program) could be executed.
Nowadays computer architectures are more complex than the single core CPU mentioned already. For instance, common architectures include those where several cores in a CPU share a common memory space and also those where CPUs are connected through some network interconnect.
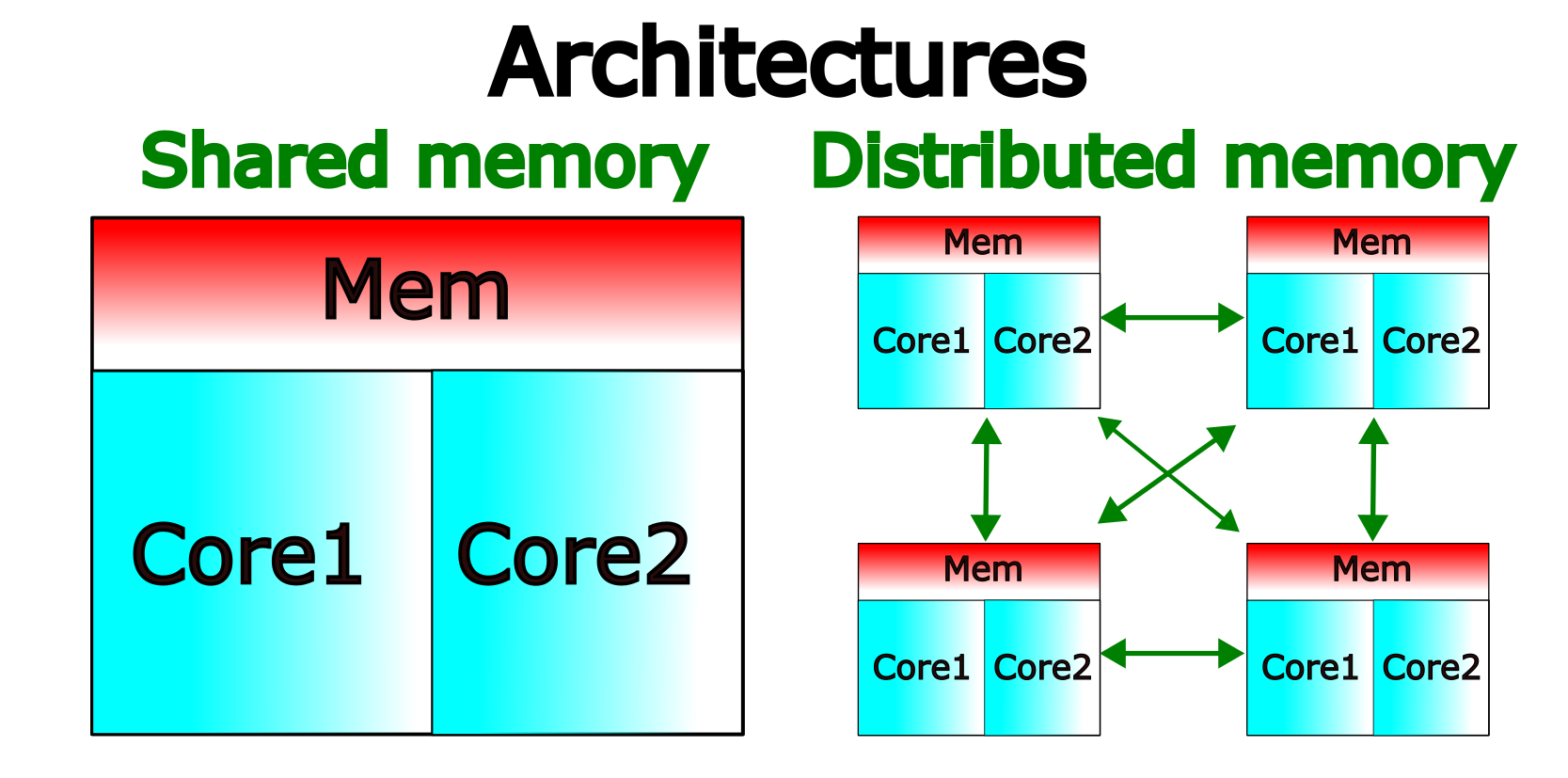
Shared Memory and Distributed Memory architectures.
A more realistic picture of a computer architecture can be seen in the following picture where we have 14 cores that shared a common memory of 64 GB. These cores form the socket and the two sockets shown in this picture constitute a node.
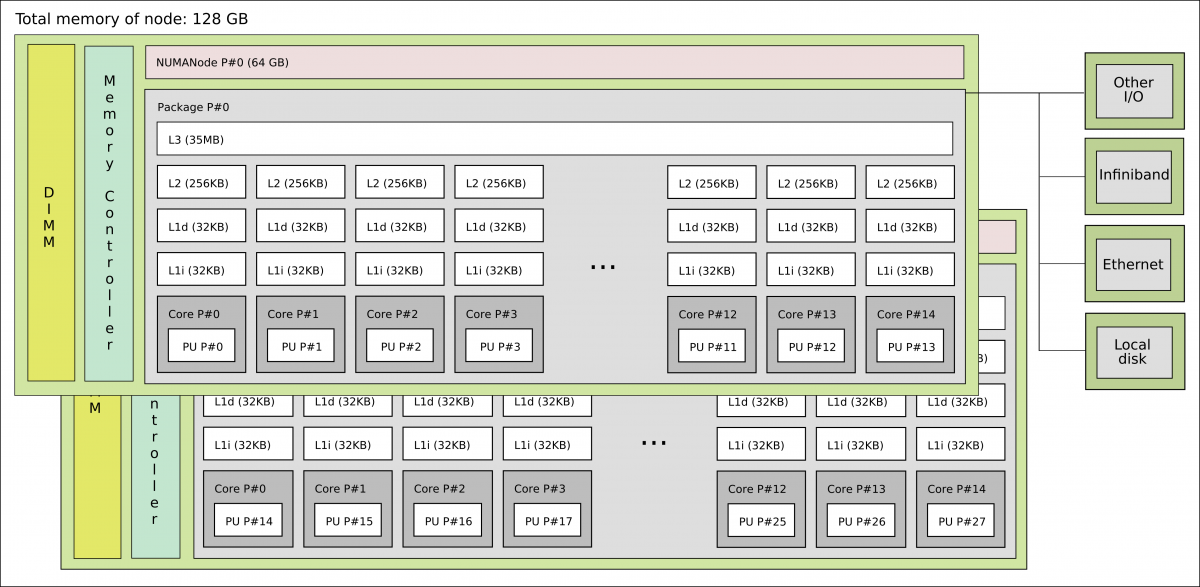
1 standard node on Kebnekaise @HPC2N
It is interesting to notice that there are different types of memory available for the cores, ranging from the L1 cache to the node’s memory for a single node. In the former, the bandwidth can be TB/s while in the latter GB/s.
Now you can see that on a single node you already have several computing units (cores) and also a hierarchy of memory resources which is denoted as Non Uniform Memory Access (NUMA).
Besides the standard CPUs, nowadays one finds Graphic Processing Units (GPUs) architectures in HPC clusters.
Why is parallel programming needed?
There is no “free lunch” when trying to use features (computing/memory resources) in modern architectures. If you want your code to be aware of those features, you will need to either add them explicitly (by coding them yourself) or implicitly (by using libraries that were coded by others).
In your local machine, you may have some number of cores available and some memory attached to them which can be exploited by using a parallel program. There can be some limited resources for running your data-production simulations as you may use your local machine for other purposes such as writing a manuscript, making a presentation, etc. One alternative to your local machine can be a High Performance Computing (HPC) cluster another could be a cloud service. A common layout for the resources in an HPC cluster is a shown in the figure below.
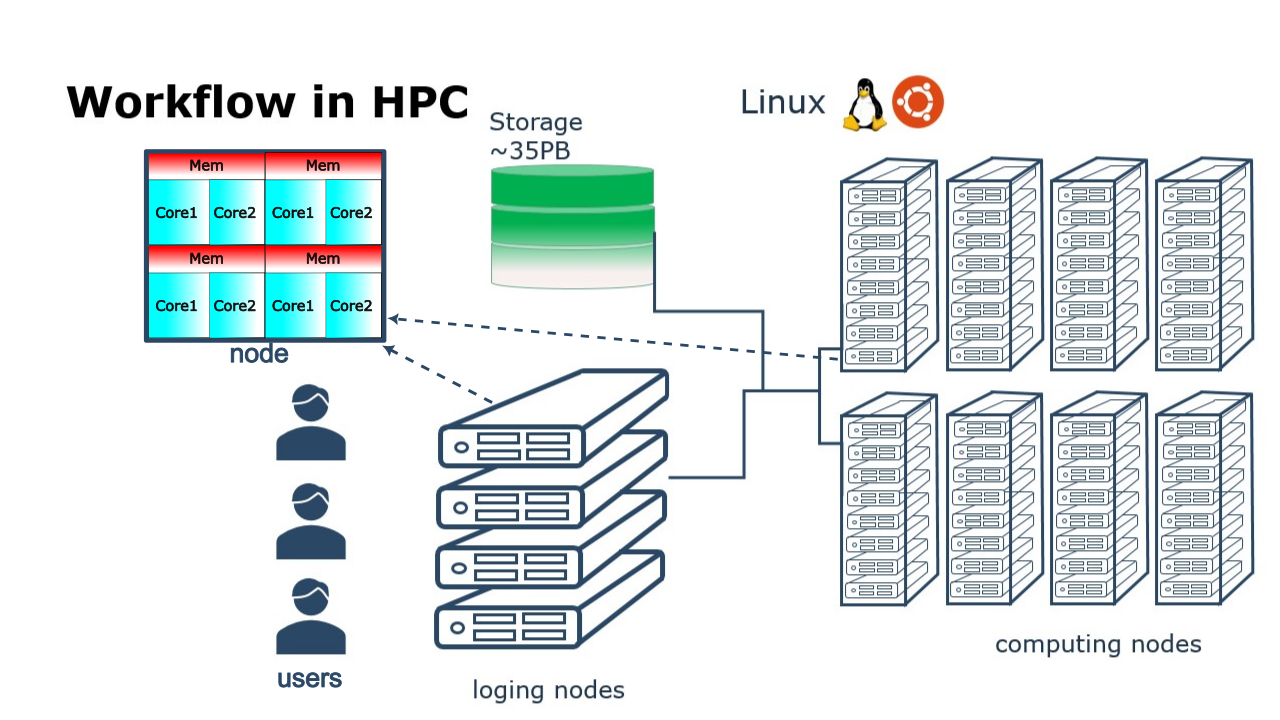
High Performance Computing (HPC) cluster.
Although a serial application can run in such a cluster, it would not gain much of the HPC resources. If fact, one can underuse the cluster if one allocates more resources than what the simulation requires.
Under-using a cluster.
Warning
Check if the resources that you allocated are being used properly.
Monitor the usage of hardware resources with tools offered at your HPC center, for instance job-usage at HPC2N.
Here there are some examples (of many) of what you will need to pay attention when porting a parallel code from your laptop (or another HPC center) to our clusters:
We have a tool to monitor the usage of resources called: job-usage at HPC2N.
If you are in a interactive node session the top command will give you information
of the resources usage.
Parallelizing code in Python
In Python there are different schemes that can be used to parallelize your code. We will only take a look at some of these schemes that illustrate the general concepts of parallel computing. The aim of this lecture is to learn how to run parallel codes in Python rather than learning to write those codes.
Demo
The idea is to parallelize a simple for loop (language-agnostic):
for i start at 1 end at 4
wait 1 second
end the for loop
The waiting step is used to simulate a task without writing too much code. In this way, one can realize how faster the loop can be executed when threads are added:
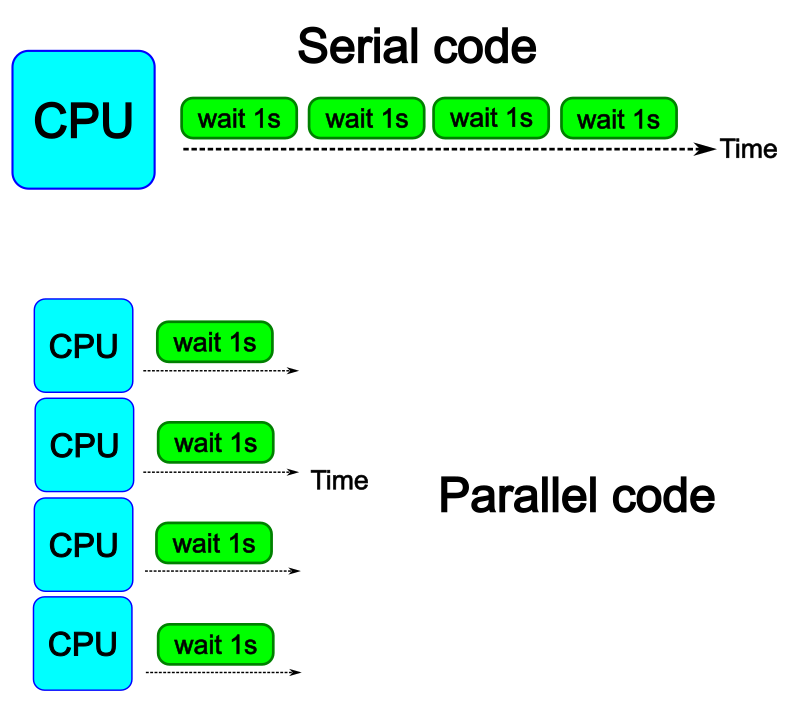
In the following example sleep.py the sleep() function is called n times first in
serial mode and then by using n processes. To parallelize the serial code we can use
the multiprocessing module that is shipped with the base library in Python so that
you don’t need to install it.
import sys
from time import perf_counter,sleep
import multiprocessing
# number of iterations
n = 4
# number of processes
numprocesses = 4
def sleep_serial(n):
for i in range(n):
sleep(1)
def sleep_threaded(n,numprocesses,processindex):
# workload for each process
workload = n/numprocesses
begin = int(workload*processindex)
end = int(workload*(processindex+1))
for i in range(begin,end):
sleep(1)
if __name__ == "__main__":
starttime = perf_counter() # Start timing serial code
sleep_serial(n)
endtime = perf_counter()
print("Time spent serial: %.2f sec" % (endtime-starttime))
starttime = perf_counter() # Start timing parallel code
processes = []
for i in range(numprocesses):
p = multiprocessing.Process(target=sleep_threaded, args=(n,numprocesses,i))
processes.append(p)
p.start()
# waiting for the processes
for p in processes:
p.join()
endtime = perf_counter()
print("Time spent parallel: %.2f sec" % (endtime-starttime))
First load the modules for Python and dependencies and then run the script
with the command srun -A "your-project" -n 1 -c 4 -t 00:05:00 python sleep.py to use 4 processes.
Optional flags for srun for writing output and error files are -o output_%j.out -e error_%j.err instead
of writing on the terminal screen.
Warning
Data dependencies
In general, one cannot parallelize a serial algorithm; let’s take the case of:
a[0] = 0
for i in range(1, N):
a[i] = a[i-1] + i
Let’s say we have the elements a[0], a[1],..., a[5] and only two processes. Process number one
takes the elements a[0], a[1], a[2] and process number two a[3], a[4], a[5].
Because in the loop the iteration i depends on the previous iteration i-1, process two will have
conflicts computing a[3] because it depends on a[2] which is owned by process one.
One can avoid data dependencies by transforming the initial algorithm to a more suitable algorithm for parallelization:
for i in range(N):
a[i] = 0.5 * i * (i + 1)
Exercises
Running a parallel code
Run the previous sleep.py code yourself using 4 processes (numprocesses = 4) and allocating 4 cores (-c 4).
In a parallel code, hardware resources can be undersubscribed (nr. cores > numprocesses) or oversubscribe (nr. cores < numprocesses). It is crucial to find the optimal number of resources.
Parallelizing a code
According to the Basel problem one can compute the summation of the reciprocals of squared integers as:
\[\sum^{\infty}_{1}\frac{1}{n^2} = \frac{1}{1^2} + \frac{1}{2^2} + \frac{1}{3^2} + \dots = \frac{\pi^2}{6} \sim 1.644934\]
Modify the previous sleep.py code (call it pi.py, for instance) to first create a serial code that computes this summation
and second a parallel version of it. To see the effects of your parallel implementation, you can run the summation up to n=1e8.
Hint
Use shared arrays of multiprocessing package to store the partial summations from each process:
from multiprocessing import Array
# number of processes
numprocesses = 4
# partial sum for each thread
partial_integrals = Array('d',[0]*numprocesses, lock=False)
In this example, the array partial_integrals has four elements partial_integrals[0],…, partial_integrals[3], that
can be accessed by each process through the processindex variable.
Solution
import sys
from time import perf_counter
import multiprocessing
from multiprocessing import Array
# number of iterations
n = int(1e8)
# number of processes
numprocesses = 4
# partial sum for each thread
partial_integrals = Array('d',[0]*numprocesses, lock=False)
def pi_serial(n):
psum = 0.0
for i in range(1,n+1):
psum = psum + 1.0/(i * i)
return psum
def pi_threaded(n,numprocesses,processindex):
# workload for each process
workload = n/numprocesses
begin = int(workload*processindex + 1)
end = int(workload*(processindex+1) + 1)
psum = 0.0
for i in range(begin,end):
psum = psum + 1.0/(i * i)
partial_integrals[processindex] = psum
if __name__ == "__main__":
starttime = perf_counter() # Start timing serial code
total_sum = pi_serial(n)
endtime = perf_counter()
print("Time spent serial: %.2f sec" % (endtime-starttime))
print("Summation = %.6f " % total_sum)
starttime = perf_counter() # Start timing parallel code
processes = []
for i in range(numprocesses):
p = multiprocessing.Process(target=pi_threaded, args=(n,numprocesses,i))
processes.append(p)
p.start()
# waiting for the processes
for p in processes:
p.join()
total_sum = sum(partial_integrals)
endtime = perf_counter()
print("Time spent parallel: %.2f sec" % (endtime-starttime))
print("Summation = %.6f " % total_sum)
2D integration
The workhorse for this section will be a 2D integration example:
One way to perform the integration is by creating a grid in the x and y directions.
More specifically, one divides the integration range in both directions into n bins. A
serial code (without any optimization) can be seen in the following code block.
integration2d_serial_initial.py
import math
import sys
from time import perf_counter
# grid size
n = 10000
if __name__ == "__main__":
starttime = perf_counter()
global integral;
# interval size (same for X and Y)
h = math.pi / float(n)
# cummulative variable
mysum = 0.0
# regular integration in the X axis
for i in range(n):
x = h * (i + 0.5)
# regular integration in the Y axis
for j in range(n):
y = h * (j + 0.5)
mysum += math.sin(x + y)
integral = h**2 * mysum
print("Integral value is %e, Error is %e" % (integral, abs(integral - 0.0)))
endtime = perf_counter()
print("Time spent: %.2f sec" % (endtime-starttime))
We can run this code on the terminal as follows:
Warning
Although this works on the terminal, having many users doing computations at the same time for this course, could create delays for other users
$ python integration2d_serial_initial.py
Integral value is -7.117752e-17, Error is 7.117752e-17
Time spent: 27.54 sec
Because of that, we can use for short-time jobs the following command:
$ srun -A <your-projec-id> -n 1 -t 00:10:00 python integration2d_serial_initial.py
Integral value is -7.117752e-17, Error is 7.117752e-17
Time spent: 26.59 sec
where srun has the flags that are used in a standard batch file.
Note that outputs can be different, when timing a code a more realistic approach
would be to run it several times to get statistics. In our initial code it is not clear if we timing the loops computation,
variable initializations, or the print() function. Thus, it is highly recommended to enclose blocks of code
(related to the same feature) into functions:
integration2d_serial.py
import math
import sys
from time import perf_counter
# grid size
n = 10000
def integration2d_serial(n):
global integral;
# interval size (same for X and Y)
h = math.pi / float(n)
# cummulative variable
mysum = 0.0
# regular integration in the X axis
for i in range(n):
x = h * (i + 0.5)
# regular integration in the Y axis
for j in range(n):
y = h * (j + 0.5)
mysum += math.sin(x + y)
integral = h**2 * mysum
if __name__ == "__main__":
starttime = perf_counter()
integration2d_serial(n)
endtime = perf_counter()
print("Integral value is %e, Error is %e" % (integral, abs(integral - 0.0)))
print("Time spent: %.2f sec" % (endtime-starttime))
Here, we can easily recognize that the part we are interested in is the one related to the nested loops computation
which is the bottleneck of the simulation. This part is now enclosed into the integration2d_serial() function.
Serial optimizations
Just before we jump into a parallelization project, Python offers some options to make
serial code faster. For instance, the Numba module can assist you to obtain a
compiled-quality function with minimal efforts. This can be achieved with the njit()
decorator:
integration2d_serial_numba.py
from numba import njit
import math
import sys
from time import perf_counter
# grid size
n = 10000
def integration2d_serial(n):
# interval size (same for X and Y)
h = math.pi / float(n)
# cummulative variable
mysum = 0.0
# regular integration in the X axis
for i in range(n):
x = h * (i + 0.5)
# regular integration in the Y axis
for j in range(n):
y = h * (j + 0.5)
mysum += math.sin(x + y)
integral = h**2 * mysum
return integral
if __name__ == "__main__":
starttime = perf_counter()
integral = njit(integration2d_serial)(n)
endtime = perf_counter()
print("Integral value is %e, Error is %e" % (integral, abs(integral - 0.0)))
print("Time spent: %.2f sec" % (endtime-starttime))
The execution time is now:
$ srun -A <your-projec-id> -n 1 -t 00:10:00 python integration2d_serial_numba.py
Integral value is -7.117752e-17, Error is 7.117752e-17
Time spent: 1.90 sec
Another option for making serial codes faster, and specially in the case of arithmetic intensive codes, is to write the most expensive parts of them in a compiled language such as Fortran or C/C++. In the next paragraphs we will show you how Fortran code for the 2D integration case can be called in Python.
We start by writing the expensive part of our Python code in a Fortran function in a file
called fortran_function.f90:
fortran_function.f90
function integration2d_fortran(n) result(integral)
implicit none
integer, parameter :: dp=selected_real_kind(15,9)
real(kind=dp), parameter :: pi=3.14159265358979323_dp
integer, intent(in) :: n
real(kind=dp) :: integral
integer :: i,j
! interval size
real(kind=dp) :: h
! x and y variables
real(kind=dp) :: x,y
! cummulative variable
real(kind=dp) :: mysum
h = pi/(1.0_dp * n)
mysum = 0.0_dp
! regular integration in the X axis
do i = 0, n-1
x = h * (i + 0.5_dp)
! regular integration in the Y axis
do j = 0, n-1
y = h * (j + 0.5_dp)
mysum = mysum + sin(x + y)
enddo
enddo
integral = h*h*mysum
end function integration2d_fortran
Then, we need to compile this code and generate the Python module (myfunction):
Warning
For UPPMAX you may have to change gcc version like:
$ ml gcc/10.3.0
Then continue…
$ f2py -c -m myfunction fortran_function.f90
running build
running config_cc
...
this will produce the Python/C API myfunction.cpython-39-x86_64-linux-gnu.so, which
can be called in Python as a module:
call_fortran_code.py
from time import perf_counter
import myfunction
import numpy
# grid size
n = 10000
if __name__ == "__main__":
starttime = perf_counter()
integral = myfunction.integration2d_fortran(n)
endtime = perf_counter()
print("Integral value is %e, Error is %e" % (integral, abs(integral - 0.0)))
print("Time spent: %.2f sec" % (endtime-starttime))
The execution time is considerably reduced:
$ srun -A <your-projec-id> -n 1 -t 00:10:00 python call_fortran_code.py
Integral value is -7.117752e-17, Error is 7.117752e-17
Time spent: 1.30 sec
Compilation of code can be tedious specially if you are in a developing phase of your code. As an alternative to improve the performance of expensive parts of your code (without using a compiled language) you can write these parts in Julia (which doesn’t require compilation) and then calling Julia code in Python. For the workhorse integration case that we are using, the Julia code can look like this:
julia_function.jl
function integration2d_julia(n::Int)
# interval size
h = π/n
# cummulative variable
mysum = 0.0
# regular integration in the X axis
for i in 0:n-1
x = h*(i+0.5)
# regular integration in the Y axis
for j in 0:n-1
y = h*(j + 0.5)
mysum = mysum + sin(x+y)
end
end
return mysum*h*h
end
A caller script for Julia would be,
call_julia_code.py
from time import perf_counter
import julia
from julia import Main
Main.include('julia_function.jl')
# grid size
n = 10000
if __name__ == "__main__":
starttime = perf_counter()
integral = Main.integration2d_julia(n)
endtime = perf_counter()
print("Integral value is %e, Error is %e" % (integral, abs(integral - 0.0)))
print("Time spent: %.2f sec" % (endtime-starttime))
from time import perf_counter
from juliacall import Main as julia
# Include the Julia script
julia.include("julia_function.jl")
# grid size
n = 10000
if __name__ == "__main__":
starttime = perf_counter()
# Call the function defined in the julia script
integral = julia.integration2d_julia(n) # function takes arguments
endtime = perf_counter()
print("Integral value is %e, Error is %e" % (integral, abs(integral - 0.0)))
print("Time spent: %.2f sec" % (endtime-starttime))
Timing in this case is similar to the Fortran serial case:
$ srun -A <your-projec-id> -n 1 -t 00:10:00 python call_julia_code.py
Integral value is -7.117752e-17, Error is 7.117752e-17
Time spent: 1.29 sec
If even with the previous (and possibly others from your own) serial optimizations your code doesn’t achieve the expected performance, you may start looking for some parallelization strategy. Here, we describe some of the Python parallel packages.
Distributed Memory
Distributed
In the distributed parallelization scheme the workers (processes) can share some common memory but they can also exchange information by sending and receiving messages for instance.
integration2d_multiprocessing.py
import multiprocessing
from multiprocessing import Array
import math
import sys
from time import perf_counter
# grid size
n = 10000
# number of processes
numprocesses = 4
# partial sum for each thread
partial_integrals = Array('d',[0]*numprocesses, lock=False)
def integration2d_multiprocessing(n,numprocesses,processindex):
global partial_integrals;
# interval size (same for X and Y)
h = math.pi / float(n)
# cummulative variable
mysum = 0.0
# workload for each process
workload = n/numprocesses
begin = int(workload*processindex)
end = int(workload*(processindex+1))
# regular integration in the X axis
for i in range(begin,end):
x = h * (i + 0.5)
# regular integration in the Y axis
for j in range(n):
y = h * (j + 0.5)
mysum += math.sin(x + y)
partial_integrals[processindex] = h**2 * mysum
if __name__ == "__main__":
starttime = perf_counter()
processes = []
for i in range(numprocesses):
p = multiprocessing.Process(target=integration2d_multiprocessing, args=(n,numprocesses,i))
processes.append(p)
p.start()
# waiting for the processes
for p in processes:
p.join()
integral = sum(partial_integrals)
endtime = perf_counter()
print("Integral value is %e, Error is %e" % (integral, abs(integral - 0.0)))
print("Time spent: %.2f sec" % (endtime-starttime))
In this case, the execution time is reduced:
$ srun -A projID -t 00:08:00 -n 1 -c 4 python integration2d_multiprocessing.py
Integral value is 4.492851e-12, Error is 4.492851e-12
Time spent: 6.06 sec
MPI
More details for the MPI parallelization scheme in Python can be found in a previous MPI course offered by some of us.
integration2d_mpi.py
from mpi4py import MPI
import math
import sys
from time import perf_counter
# MPI communicator
comm = MPI.COMM_WORLD
# MPI size of communicator
numprocs = comm.Get_size()
# MPI rank of each process
myrank = comm.Get_rank()
# grid size
n = 10000
def integration2d_mpi(n,numprocs,myrank):
# interval size (same for X and Y)
h = math.pi / float(n)
# cummulative variable
mysum = 0.0
# workload for each process
workload = n/numprocs
begin = int(workload*myrank)
end = int(workload*(myrank+1))
# regular integration in the X axis
for i in range(begin,end):
x = h * (i + 0.5)
# regular integration in the Y axis
for j in range(n):
y = h * (j + 0.5)
mysum += math.sin(x + y)
partial_integrals = h**2 * mysum
return partial_integrals
if __name__ == "__main__":
starttime = perf_counter()
p = integration2d_mpi(n,numprocs,myrank)
# MPI reduction
integral = comm.reduce(p, op=MPI.SUM, root=0)
endtime = perf_counter()
if myrank == 0:
print("Integral value is %e, Error is %e" % (integral, abs(integral - 0.0)))
print("Time spent: %.2f sec" % (endtime-starttime))
Execution of this code gives the following output:
$ mpirun -np 4 python integration2d_mpi.py
Integral value is 4.492851e-12, Error is 4.492851e-12
Time spent: 5.76 sec
For long jobs or MPI jobs, one will need to run in batch mode. Here is an example of a batch script for this MPI example,
#!/bin/bash -l
#SBATCH -A naiss202X-XY-XYZ
#SBATCH -t 00:05:00
#SBATCH -n 4
#SBATCH -o output_%j.out # output file
#SBATCH -e error_%j.err # error messages
ml buildenv-gcccuda/12.2.2-gcc11-hpc1
#ml julia/1.10.2-bdist # if Julia is needed
source /path-to-your-project/vpyenv-python-course/bin/activate
mpirun -np 4 python integration2d_mpi.py
#!/bin/bash
#SBATCH -A hpc2n-20XX-XYZ
#SBATCH -t 00:05:00 # wall time
#SBATCH -n 4
#SBATCH -o output_%j.out # output file
#SBATCH -e error_%j.err # error messages
ml purge > /dev/null 2>&1
ml GCCcore/11.2.0 Python/3.9.6
ml GCC/11.2.0 OpenMPI/4.1.1
#ml Julia/1.7.1-linux-x86_64 # if Julia is needed
source /proj/nobackup/<your-project-storage>/vpyenv-python-course/bin/activate
mpirun -np 4 python integration2d_mpi.py
#!/bin/bash -l
#SBATCH -A naiss-202X-XY-XYZ
#SBATCH -t 00:05:00
#SBATCH -n 4
#SBATCH -o output_%j.out # output file
#SBATCH -e error_%j.err # error messages
ml python/3.9.5
ml gcc/9.3.0 openmpi/3.1.5
#ml julia/1.7.2 # if Julia is needed
source /proj/naiss202X-XY-XYZ/nobackup/<user>/venv-python-course/bin/activate
mpirun -np 4 python integration2d_mpi.py
#!/bin/bash
#SBATCH -A lu202u-vw-xy
#SBATCH -t 00:05:00
#SBATCH -n 4
#SBATCH -o output_%j.out # output file
#SBATCH -e error_%j.err # error messages
ml GCC/12.3.0 Python/3.11.3 OpenMPI/4.1.5
#ml Julia/1.10.4-linux-x86_64 # if Julia is needed
source /path-to-your-project/vpyenv-python-course/bin/activate
mpirun -np 4 python integration2d_mpi.py
#!/bin/bash
#SBATCH -A naiss202t-uv-wxyz
#SBATCH -t 00:05:00
#SBATCH -p shared # name of the queue
#SBATCH --ntasks=4 # nr. of tasks
#SBATCH --cpus-per-task=1 # nr. of cores per-task
#SBATCH -o output_%j.out # output file
#SBATCH -e error_%j.err # error messages
ml cray-python
source /path-to-your-project/vpyenv-python-course/bin/activate
srun python integration2d_mpi.py
Heat (Advanced)
Heat is a library for distributing tensor operations by using MPI as a backend. Heat uses Distributed N-Dimensional (DND) arrays that can be seen as a global array. Locally, each rank retains a chunk of the array which is a tensor PyTorch tensor:
heat_datatypes.py
import heat as ht
x = ht.arange(10, split=0)
print(type(x)) # <class 'heat.core.dndarray.DNDarray'>
print(x.shape) # <class 'heat.core.dndarray.DNDarray'>
print(type(x.larray)) # <class 'torch.Tensor'>
print(x.larray.shape) # <class 'torch.Tensor'>
It is recommended to use a batch script for Heat scripts:
#!/bin/bash -l
#SBATCH -A naiss202X-XY-XYZ
#SBATCH -t 00:05:00
#SBATCH -n 1
#SBATCH -c 32
#SBATCH --gpus-per-task=1
#SBATCH -o output_%j.out # output file
#SBATCH -e error_%j.err # error messages
ml buildenv-gcccuda/12.2.2-gcc11-hpc1
#ml julia/1.10.2-bdist # if Julia is needed
source /path-to-your-project/vpyenv-python-course/bin/activate
mpirun --oversubscribe -np 2 python heat_datatypes.py
#!/bin/bash
#SBATCH -A hpc2n-20XX-XYZ
#SBATCH -t 00:05:00 # wall time
#SBATCH -n 2
#SBATCH -o output_%j.out # output file
#SBATCH -e error_%j.err # error messages
ml purge > /dev/null 2>&1
ml GCCcore/11.2.0 Python/3.9.6
ml GCC/11.2.0 OpenMPI/4.1.1
#ml Julia/1.7.1-linux-x86_64 # if Julia is needed
source /proj/nobackup/<your-project-storage>/vpyenv-python-course/bin/activate
mpirun -np 2 python heat_datatypes.py
On Kebnekaise, the srun command also works:
$ srun -A projectID -t 00:08:00 -n 2 python heat_datatypes.py
Output:
(10,)
<class 'torch.Tensor'>
torch.Size([5])
<class 'heat.core.dndarray.DNDarray'>
(10,)
<class 'torch.Tensor'>
torch.Size([5])
More details for this package can be found here Heat package.
Example 1: Distributing matrix-matrix multiplication operations
heat_matmat.py
import heat as ht
import torch
import time
if torch.cuda.is_available():
device = "gpu"
else:
device = "cpu"
# load large matrices distributed across CPUs/GPUs
A = ht.random.randn(10000, 10000, split=0, device=device) # Split along rows
B = ht.random.randn(10000, 10000, split=None, device=device) # Do not split just copy
# Perform distributed matrix multiplication
start = time.time()
C = ht.linalg.matmul(A, B)
end = time.time()
print("rank= ", ht.MPI_WORLD.rank, "reported time= ", end-start)
Example 2: Distributing gradients
heat_gradients.py
import heat as ht
import torch
import torch.nn as nn
import torch.optim as optim
import time
# Distributed dataset
X = ht.random.randn(30000, 10000, split=0)
y = ht.random.randn(30000, 1, split=0)
# Local model per rank
model = nn.Linear(10000, 1)
optimizer = optim.SGD(model.parameters(), lr=0.01)
start = time.time()
# Train local arrays
for epoch in range(1000):
out = model(X.larray)
loss = torch.mean((out - y.larray) ** 2)
optimizer.zero_grad()
loss.backward()
optimizer.step()
# Average parameters across ranks (distributed synchronization)
for param in model.parameters():
ht.MPI_WORLD.Allreduce(ht.communication.MPI.IN_PLACE, param.data, op=ht.communication.MPI.SUM)
param.data /= ht.MPI_WORLD.size
end = time.time()
print("rank= ", ht.MPI_WORLD.rank, "reported time= ", end-start)
print(f"Rank {ht.MPI_WORLD.rank} done training")
Monitoring resources’ usage
Monitoring the resources that a certain job uses is important specially when this job is expected to run on many CPUs and/or GPUs. It could happen, for instance, that an incorrect module is loaded or the command for running on many CPUs is not the proper one and our job runs in serial mode while we allocated possibly many CPUs/GPUs. For this reason, there are several tools available in our centers to monitor the performance of running jobs.
HPC2N
On a Kebnekaise terminal, you can type the command:
$ job-usage job_ID
where job_ID is the number obtained when you submit your job with the sbatch
command. This will give you a URL that you can copy and then paste in your local
browser. The results can be seen in a graphical manner a couple of minutes after the
job starts running, here there is one example of how this looks like:
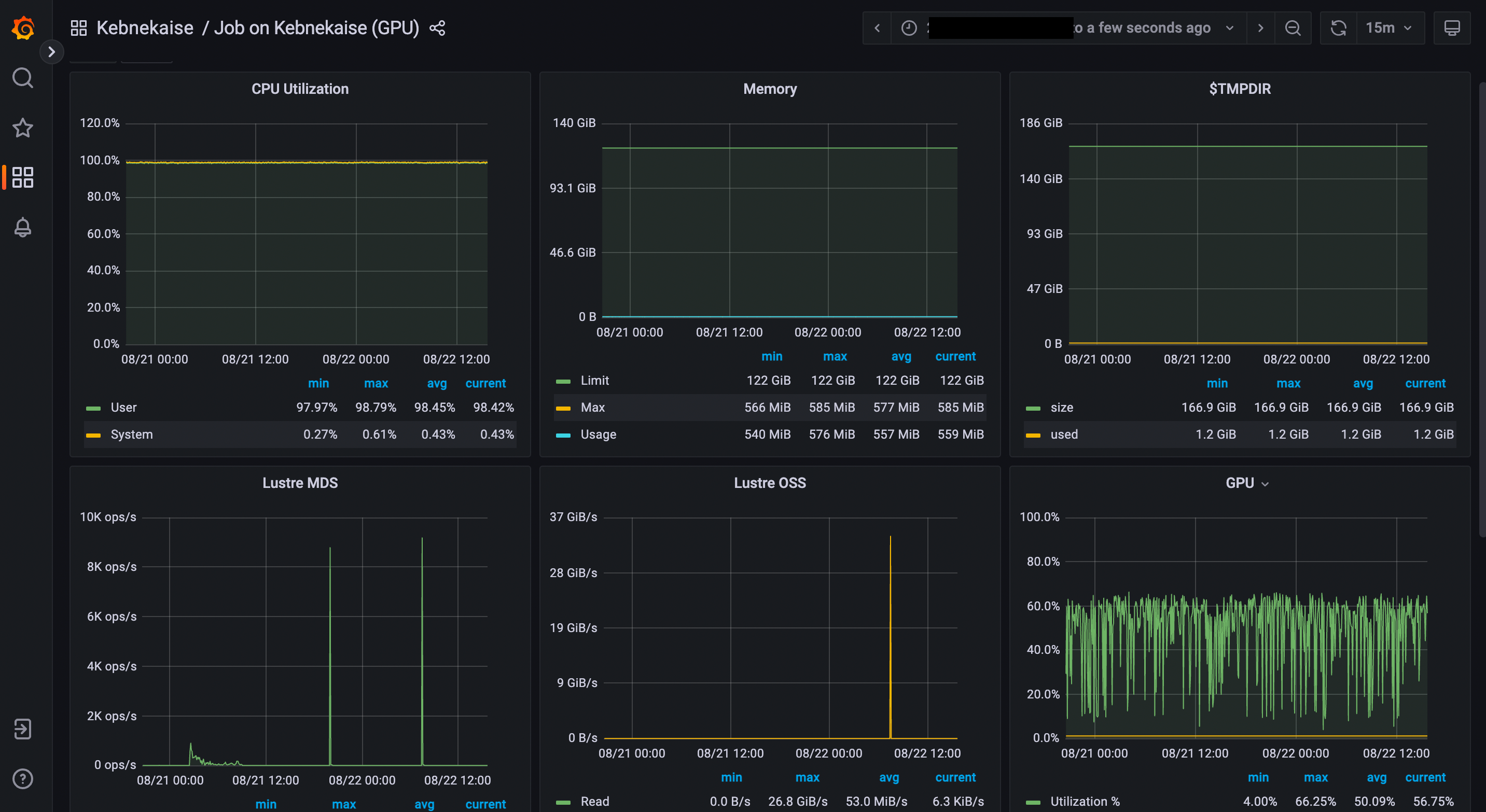
The resources used by a job can be monitored in your local browser. For this job, we can notice that 100% of the requested CPU and 60% of the GPU resources are being used.
Exercises
Running a parallel code efficiently
In this exercise we will run the parallelized code that performs a 2D integration:
using the multiprocessing module in Python that we described above:
integration2d_multiprocessing.py
import multiprocessing
from multiprocessing import Array
import math
import sys
from time import perf_counter
# grid size
n = 5000
# number of processes
numprocesses = *FIXME*
# partial sum for each thread
partial_integrals = Array('d',[0]*numprocesses, lock=False)
# Implementation of the 2D integration function (non-optimal implementation)
def integration2d_multiprocessing(n,numprocesses,processindex):
global partial_integrals;
# interval size (same for X and Y)
h = math.pi / float(n)
# cummulative variable
mysum = 0.0
# workload for each process
workload = n/numprocesses
begin = int(workload*processindex)
end = int(workload*(processindex+1))
# regular integration in the X axis
for i in range(begin,end):
x = h * (i + 0.5)
# regular integration in the Y axis
for j in range(n):
y = h * (j + 0.5)
mysum += math.sin(x + y)
partial_integrals[processindex] = h**2 * mysum
if __name__ == "__main__":
starttime = perf_counter()
processes = []
for i in range(numprocesses):
p = multiprocessing.Process(target=integration2d_multiprocessing, args=(n,numprocesses,i))
processes.append(p)
p.start()
# waiting for the processes
for p in processes:
p.join()
integral = sum(partial_integrals)
endtime = perf_counter()
print("Integral value is %e, Error is %e" % (integral, abs(integral - 0.0)))
print("Time spent: %.2f sec" % (endtime-starttime))
Run the code with the following batch script:
job.sh
#!/bin/bash -l
#SBATCH -A naiss202X-XY-XYZ # your project_ID
#SBATCH -J job-serial # name of the job
#SBATCH -N 1
#SBATCH -c *FIXME* # nr. coresw
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
# Load any required modules
ml buildenv-gcccuda/12.2.2-gcc11-hpc1
python integration2d_multiprocessing.py
#!/bin/bash -l
#SBATCH -A naiss202X-XY-XYZ # your project_ID
#SBATCH -J job-serial # name of the job
#SBATCH -n *FIXME* # nr. tasks/coresw
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
# Load any modules you need, here for Python 3.11.8 and compatible SciPy-bundle
module load python/3.11.8
python integration2d_multiprocessing.py
#!/bin/bash
#SBATCH -A hpc2n202X-XYZ # your project_ID
#SBATCH -J job-serial # name of the job
#SBATCH -n *FIXME* # nr. tasks
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
# Do a purge and load any modules you need, here for Python
ml purge > /dev/null 2>&1
ml GCCcore/11.2.0 Python/3.9.6
python integration2d_multiprocessing.py
#!/bin/bash
#SBATCH -A lu202X-XX-XX # your project_ID
#SBATCH -J job-serial # name of the job
#SBATCH -n *FIXME* # nr. tasks
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
# reservation (optional)
#SBATCH --reservation=RPJM-course*FIXME*
# Do a purge and load any modules you need, here for Python
ml purge > /dev/null 2>&1
ml GCCcore/12.3.0 Python/3.11.3
python integration2d_multiprocessing.py
#!/bin/bash -l
#SBATCH -A naiss202X-XY-XYZ # your project_ID
#SBATCH -J job-serial # name of the job
#SBATCH -p shared # name of the queue
#SBATCH --ntasks=*FIXME* # nr. of tasks
#SBATCH --cpus-per-task=1 # nr. of cores per-task
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
# Load Python
ml cray-python
python integration2d_multiprocessing.py
Try different number of cores for this batch script (FIXME string) using the sequence: 1,2,4,8,12, and 14. Note: this number should match the number of processes (also a FIXME string) in the Python script. Collect the timings that are printed out in the job.*.out. According to these execution times what would be the number of cores that gives the optimal (fastest) simulation?
Challenge: Increase the grid size (n) to 15000 and submit the batch job with 4 workers (in the
Python script) and request 5 cores in the batch script. Monitor the usage of resources
with tools available at your center, for instance top (UPPMAX) or
job-usage (HPC2N).
Parallelizing a for loop workflow (Advanced)
Create a Data Frame containing two features, one called ID which has integer values
from 1 to 10000, and the other called Value that contains 10000 integers starting from 3
and goes in steps of 2 (3, 5, 7, …). The following codes contain parallelized workflows
whose goal is to compute the average of the whole feature Value using some number of
workers. Substitute the FIXME strings in the following codes to perform the tasks given
in the comments. Call the script for instance script-df.py.
Warning
For Tetralith you will need to install pandas:
ml buildenv-gcccuda/12.2.2-gcc11-hpc1
source vpyenv-python-course/bin/activate
pip install pandas
Pandas is available in the following combo ml GCC/12.3.0 SciPy-bundle/2023.07 (HPC2N) and
ml python/3.11.8 (UPPMAX).
import pandas as pd
import multiprocessing
# Create a DataFrame with two sets of values ID and Value
data_df = pd.DataFrame({
'ID': range(1, 10001),
'Value': range(3, 20002, 2) # Generate 10000 odd numbers starting from 3
})
# Define a function to calculate the sum of a vector
def calculate_sum(values):
total_sum = *FIXME*(values)
return *FIXME*
# Split the 'Value' column into chunks of size 1000
chunk_size = *FIXME*
value_chunks = [data_df['Value'][*FIXME*:*FIXME*] for i in range(0, len(data_df['*FIXME*']), *FIXME*)]
# Create a Pool of 4 worker processes, this is required by multiprocessing
pool = multiprocessing.Pool(processes=*FIXME*)
# Map the calculate_sum function to each chunk of data in parallel
results = pool.map(*FIXME: function*, *FIXME: chunk size*)
# Close the pool to free up resources, if the pool won't be used further
pool.close()
# Combine the partial results to get the total sum
total_sum = sum(results)
# Compute the mean by dividing the total sum by the total length of the column 'Value'
mean_value = *FIXME* / len(data_df['*FIXME*'])
# Print the mean value
print(mean_value)
Run the code with the batch script:
#!/bin/bash -l
#SBATCH -A naiss202X-XY-XYZ # your project_ID
#SBATCH -J job-serial # name of the job
#SBATCH -n 4 # nr. tasks/coresw
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
# Load any required modules
ml buildenv-gcccuda/12.2.2-gcc11-hpc1
source vpyenv-python-course/bin/activate
python script-df.py
#!/bin/bash -l
#SBATCH -A naiss202u-w-xyz # your project_ID
#SBATCH -J job-parallel # name of the job
#SBATCH -n 4 # nr. tasks/coresw
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
# Load any modules you need, here for Python 3.11.8 and compatible SciPy-bundle
module load python/3.11.8
python script-df.py
#!/bin/bash
#SBATCH -A hpc2n202w-xyz # your project_ID
#SBATCH -J job-parallel # name of the job
#SBATCH -n 4 # nr. tasks
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
# Load any modules you need, here for Python 3.11.3 and compatible SciPy-bundle
module load GCC/12.3.0 Python/3.11.3 SciPy-bundle/2023.07
python script-df.py
#!/bin/bash
#SBATCH -A lu202u-vw-xyz # your project_ID
#SBATCH -J job-parallel # name of the job
#SBATCH -n 4 # nr. tasks
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
#SBATCH --reservation=RPJM-course*FIXME* # reservation (optional)
# Purge and load any modules you need, here for Python & SciPy-bundle
ml purge
ml GCCcore/12.3.0 Python/3.11.3 SciPy-bundle/2023.07
python script-df.py
#!/bin/bash -l
#SBATCH -A naiss202u-vw-xyz # your project_ID
#SBATCH -J job-parallel # name of the job
#SBATCH -p shared # name of the queue
#SBATCH --ntasks=4 # nr. of tasks
#SBATCH --cpus-per-task=1 # nr. of cores per-task
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
# Load any modules you need, here for Python 3.11.8 and compatible SciPy-bundle
module load cray-python
python script-df.py
Solution
import pandas as pd
import multiprocessing
# Create a DataFrame with two sets of values ID and Value
data_df = pd.DataFrame({
'ID': range(1, 10001),
'Value': range(3, 20002, 2) # Generate 10000 odd numbers starting from 3
})
# Define a function to calculate the sum of a vector
def calculate_sum(values):
total_sum = sum(values)
return total_sum
# Split the 'Value' column into chunks
chunk_size = 1000
value_chunks = [data_df['Value'][i:i+chunk_size] for i in range(0, len(data_df['Value']), chunk_size)]
# Create a Pool of 4 worker processes, this is required by multiprocessing
pool = multiprocessing.Pool(processes=4)
# Map the calculate_sum function to each chunk of data in parallel
results = pool.map(calculate_sum, value_chunks)
# Close the pool to free up resources, if the pool won't be used further
pool.close()
# Combine the partial results to get the total sum
total_sum = sum(results)
# Compute the mean by dividing the total sum by the total length of the column 'Value'
mean_value = total_sum / len(data_df['Value'])
# Print the mean value
print(mean_value)
See also
The book High Performance Python is a good resource for ways of speeding up Python code.
Keypoints
You deploy cores and nodes via SLURM, either in interactive mode or batch
In Python, threads, distributed and MPI parallelization can be used.