Using IDEs
There are several popular IDEs that are commonly used for interactive work with Python. Here we will show how to load Jupyter, VS Code, and Spyder.
Jupyter
JupyterLab 🚀
JupyterLab is the next-generation web-based user interface for Project Jupyter. It provides an interactive development environment for working with notebooks, code, and data. JupyterLab offers a more flexible and powerful interface compared to the classic Jupyter Notebook, allowing users to arrange multiple documents and activities side by side in tabs or split screens.
Jupyter Notebook 📝
Jupyter Notebook is a sibling to other notebook authoring applications under the Project Jupyter umbrella, like JupyterLab. Jupyter Notebook offers a lightweight, simplified experience compared to JupyterLab.
Running python requires a kernel to be started. IPython is the default kernel for Jupyter if you want to run python code. You can also install other kernels to run other programming languages.
A Jupyter Notebook is made up of cells. There are two main types of cells: Code cells and Markdown cells.
Code cells 🧩 allow you to write and execute Python code. When you run a code cell, the output is displayed directly below the cell.
Markdown cells 📝 allow you to write formatted text using Markdown syntax. This is useful for adding explanations, headings, lists, links, and even LaTeX equations to your notebook.
You can run a cell by selecting it and pressing
Shift + Enteror by clicking the “Run” button in the toolbar ▶️.Cell execution order matters - Jupyter allows cells to be run in any order, but the execution number in square brackets shows the order in which cells were actually run.
Restarting the kernel 🔄 is useful when you want to clear all variables and start fresh. Go to “Kernel” > “Restart”.
Clearing all outputs 🧹 can be done by selecting “Cell” > “All Output” > “Clear” from the menu.
Exporting notebooks 📤 to different formats (HTML, PDF, Markdown, etc.) can be done via “File” > “Download as” or “Export Notebook As”.
Using magic commands ✨ like
%timeit,%matplotlib inline,%load, and%%writefilecan enhance your workflow significantly.
Keyboard shortcuts
Shift + Enter: Run the current cell and move to the next oneCtrl + Enter: Run the current cell and stay in itAlt + Enter: Run the current cell and insert a new one belowEsc: Enter command modeEnter: Enter edit mode- In command mode:
A: Insert cell aboveB: Insert cell belowDD: Delete current cellM: Change cell to MarkdownY: Change cell to CodeShift + Up/Down: Select multiple cells
Magic commands
Use these to speed up development, debugging, and exploration inside notebooks:
%lsmagic— list available line and cell magics.%lsmagic
- Timing snippets
%time— time a single statement.
%time sum(range(100000))
%timeit/%%timeit— run repeated timings for more robust measurements.
%timeit sum(range(1000)) %%timeit a = [i*i for i in range(1000)] sum(a)
%run— run a Python script and load its variables into the notebook namespace.%run scripts/my_analysis.py
%load— load a script or URL into the current cell (useful for editing before execution).%load scripts/snippet.py
- Auto-reload for iterative development
%load_ext autoreload %autoreload 2 # Now imported modules are reloaded automatically when changed.
- Variable inspection and namespace control
%who # names only %whos # detailed table of variables %reset -f # clear user namespace
- Interactive debugger on exceptions
%pdb on # Raises into the interactive debugger when exceptions occur
- Persist variables between sessions
mydata = {"a": 1} %store mydata # In a later session: %store -r mydata
- Installing packages from within the notebook (preferred over !pip)
%pip install matplotlib seaborn
- Run shell blocks or single shell commands
%%bash echo "This runs in a bash subshell" ls -l # single-line shell: !pwd
- IPython profiling and line profiling (if extensions are installed)
%prun my_function() # run profiler %lprun -f my_function my_function() # requires line_profiler extension
- Show available magics and help
%magic # detailed magic help %timeit? # doc for a specific magic
Exercise
Try Jupyter interface with the following Notebook Code.
Either start an interactive session with 4-8 cores and 1-2 hr walltime or stay on the login node.
cd into your project directory and start Jupyter Notebook or JupyterLab as described in previous sections. (Optional: load matplotlib module by searching it with module spider matplotlib and then loading it.)
Create a new Python 3 notebook.
Notebook Code
Type out (or copy) the following code snippets (Cell 1, Cell 2,…) into different cells in a Jupyter Notebook and run them to see the results.
If there is a missing package while running the following examples, dont worry, those will be covered in later sessions.
# Cell 1: Basic arithmetic and print
a = 10
b = 20
result = a + b
print(f"The sum of {a} and {b} is: {result}")
# Cell 2: Markdown example
# Run this in markdown mode:
## This is a heading
This is **bold** text and this is *italic* text.
- List item 1
- List item 2
# LaTeX math equation:
$$E = mc^2$$
or inline: $a^2 + b^2 = c^2$
# Cell 3: Using magic commands
# Check execution time
%timeit sum(range(1000))
# Cell 4: List all variables
%whos
# Cell 5: Run system commands
!pwd
!ls -l
# Cell 6: Display plot inline
%matplotlib inline
import matplotlib.pyplot as plt
x = [1, 2, 3, 4, 5]
y = [1, 4, 9, 16, 25]
plt.plot(x, y)
plt.title("Simple Plot")
plt.show()
Resources
Official Jupyter documentation: Jupyter Documentation
Jupyter notebook examples from original documentation
You can also check the lesson about Jupyter on compute nodes in our “Introduction to running R, Python and Julia in HPC workshop”
Documentation about Jupyter on HPC2N
Documentation about Jupyter on UPPMAX
Documentation about Jupyter on C3SE
Documentation about Jupyter on LUNARC
Spyder
Features
When you open Spyder you will see a three-pane layout similar to other scientific IDEs: a large Editor on the left, and one or more utility panes on the right and bottom. The exact arrangement can be customised and saved as a layout.
Spyder interface
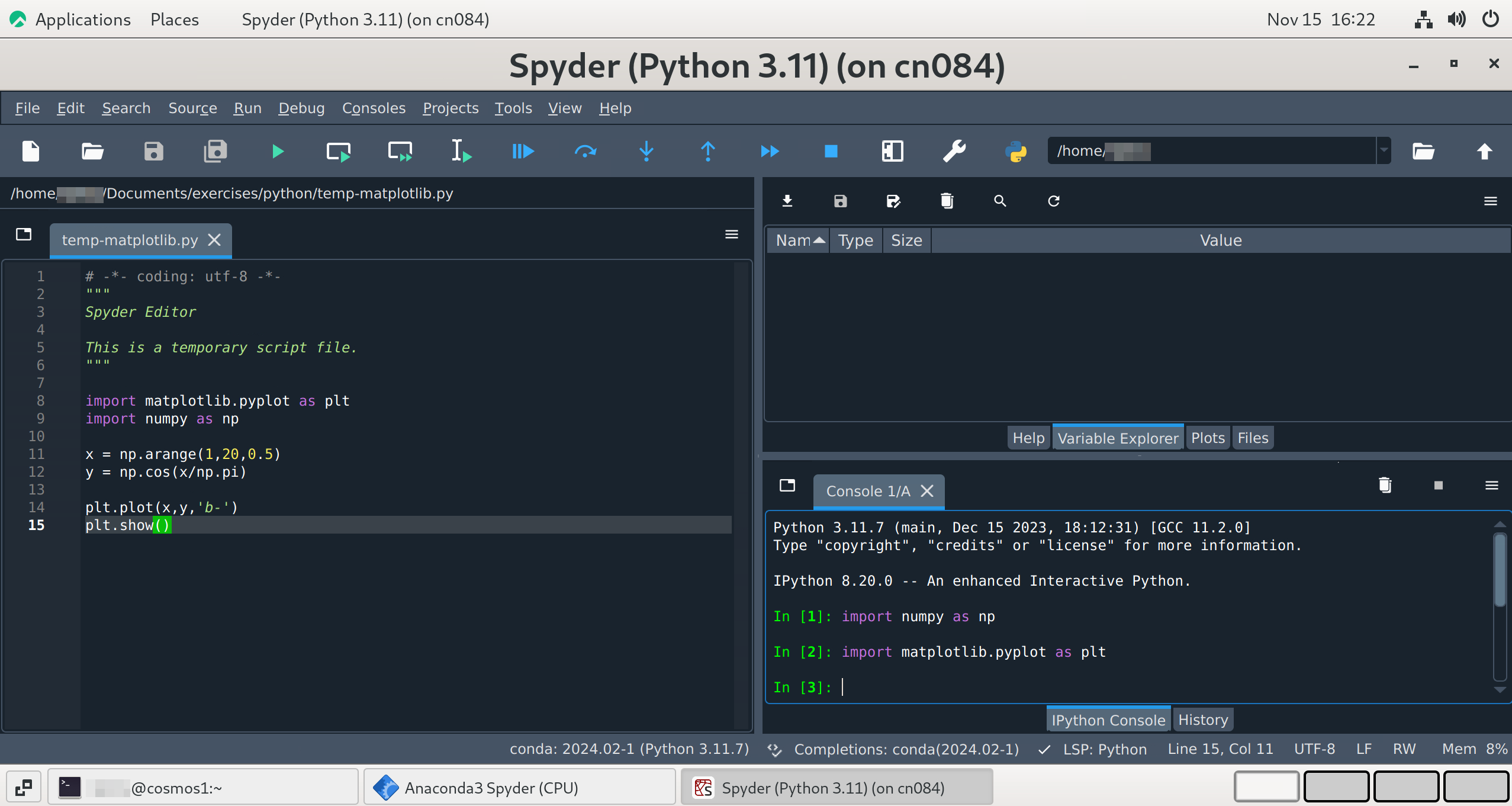
Main panes and useful tabs
- Editor (left) 📝
Edit multiple files in tabs : use the Run toolbar (green ▶️) to run the current file or selection.
You can mark cells with
# %%(or# %%%for multi level cells) and run cells interactively.Use the burger menu on the editor pane to split, undock or save layout.
- Utility pane (Top-right) 🧰
Help 🔍: documentation and contextual help for the symbol under the cursor.
Variable explorer 🧾 : table view of in-memory variables with types and values; inspect, edit or remove variables.
Files 📂 : file browser for the current working directory.
Plots 📈 : interactive plots panel (may be a tab in recent versions); can be undocked to a separate window for better interaction.
- Console and History (Bottom-right) 🖥️
IPython console(s) : interactive Python connected to the currently selected kernel/environment.
History log : previously executed commands from the console.
Quick Tips
- Run workflow
Save your script and click the Run button to execute it in the IPython console. Running a saved file will switch the console working directory to the file’s location (configurable in Preferences).
Run selection or a cell from the editor to test snippets without running the whole file.
- Debugging
Set breakpoints in the editor gutter and launch the debugger from the Run menu or toolbar to step through code and inspect variables.
- Plotting
Undock the Plots tab or set the IPython console graphics backend to “Automatic” or a GUI backend in Preferences so figures open in separate windows and remain interactive.
- Multiple consoles/kernels
You can open multiple IPython consoles and assign each a different interpreter or conda environment. This is useful for testing code against different environments.
- Notes
Undock panels (burger menu → Undock) for a multi-monitor workflow or to resize plots independently.
If Spyder appears too small on ThinLinc or high-DPI screens, enable “Set custom high-DPI scaling” in Preferences → General → Interface.
Prefer launching Spyder from a graphical session (ThinLinc / On-demand desktop). Running it on a login node without a desktop may not work or be unsupported.
Configuring Spyder
Font and icon sizes. If you are on Thinlinc and/or on a Linux machine, Spyder may be uncomfortably small the first time you open it. To fix this,
Click the icon shaped like a wrench (Preferences) or click “Tools” and then “Preferences”. A popup should open with a menu down the left side, and the “General” menu option should already be selected (if not, select it now).
You should see a tab titled “Interface” that has options that mention “high-DPI scaling”. Select “Set custom high-DPI scaling” and enter the factor by which you’d like the text and icons to be magnified (recommend a number from 1.5 to 2).
Click “Apply”. If the popup in the next step doesn’t appear immediately, then click “OK”.
A pop-up will appear that says you need to restart to view the changes and asks if you want to restart now. Click “Yes” and wait. The terminal may flash some messages like
QProcess: Destroyed while process ("/hpc2n/eb/software/Python/3.8.2-GCCcore-9.3.0/bin/python") is still running.", but it should restart within a minute or so. Don’t interrupt it or you’ll have to start over.
The text and icons should be rescaled when it reopens, and should stay rescaled even if you close and reopen Spyder, as long as you’re working in the same session.
(Optional but recommended) Configure plots to open in separate windows. In some versions of Spyder, there is a separate Plotting Pane that you can click and drag out of its dock so you can resize figures as needed, but if you don’t see that, you will probably want to change your graphics backend. The default is usually “Inline”, which is usually too small and not interactive. To change that,
Click the icon shaped like a wrench (Preferences) or click “Tools” and then “Preferences” to open the Preferences popup.
In the menu sidebar to the left, click “IPython console”. The box to the right should then have 4 tabs, of which the second from the left is “Graphics” (see figure below).
Click “Graphics” and find the “Graphics backend” box below. In that box, next to “Backend” there will be a dropdown menu that probably says “Inline”. Click the dropdown and select “Automatic”.
Click “Apply” and then “OK” to exit.
Spyder interface
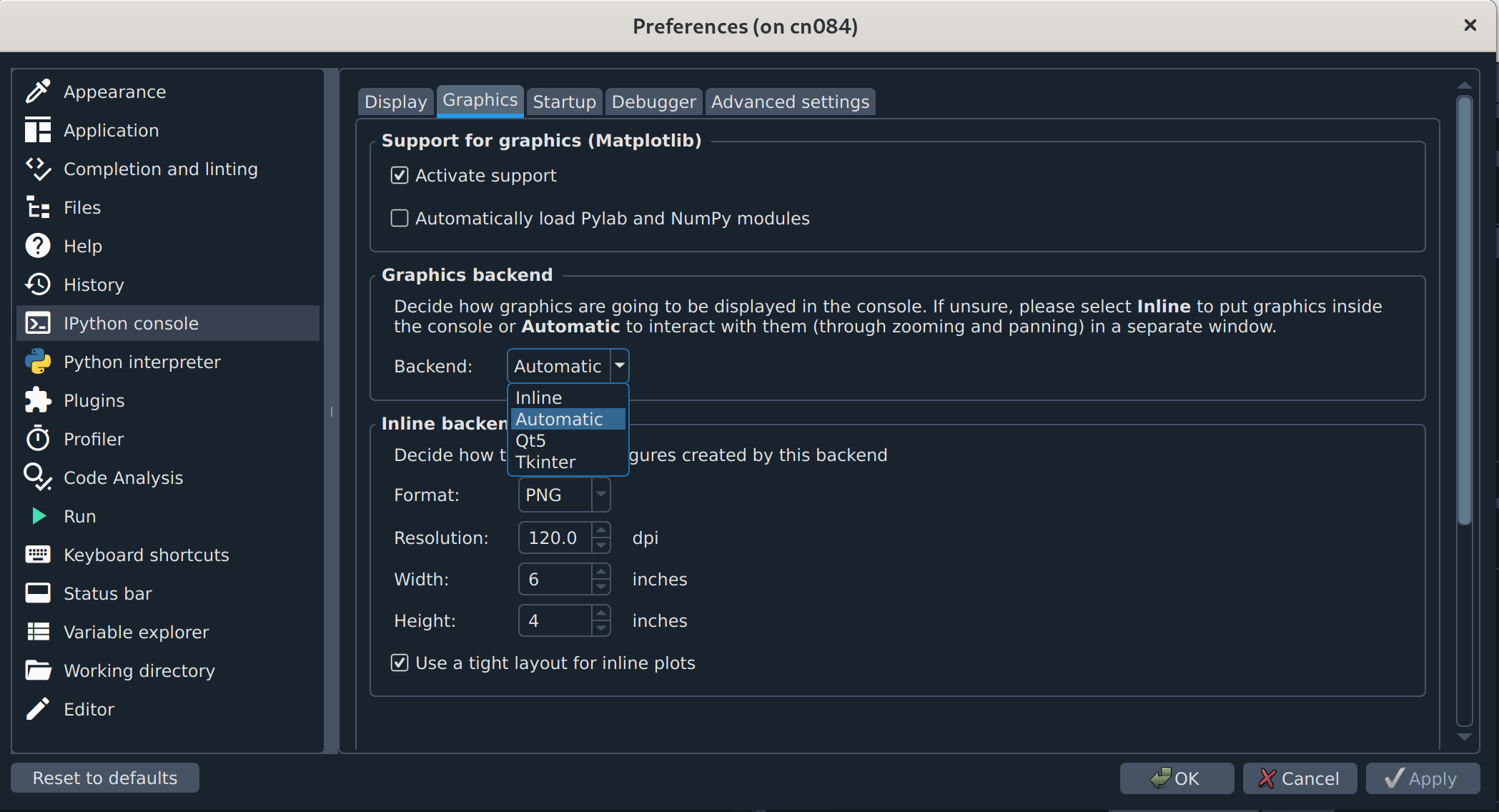
Now, graphics should appear in their own popup that has menu options to edit and save the content.
Exercise
Try Spyder interface with the following code.
Either start an interactive session with 4-8 cores and 1-2 hr walltime or stay on the login node.
cd into your project directory.
Start Spyder from your python env that you created in previous session.
Type out (or copy) the following code into the IDE editor and run it to see the results.
spyder_basics.py
# %% Basic variables — explore these in the Variable Explorer
a = 10
b = 3.5
c = a * b
message = "Hello Spyder"
data = list(range(10))
matrix = [[i * j for j in range(5)] for i in range(3)]
print(message)
# %% Class and state — inspect and modify instances from the Variable Explorer
class Counter:
"""Simple counter object to experiment with the Variable Explorer."""
def __init__(self, start=0):
self.value = start
def inc(self, n=1):
self.value += n
def reset(self):
self.value = 0
def __repr__(self):
return f"Counter({self.value})"
ctr = Counter(5)
ctr.inc(2)
# A small dict to glance at many variable types in the Variable Explorer
__all_vars__ = dict(
a=a, b=b, c=c, message=message, data=data,
matrix=matrix, ctr=ctr
)
# %% Run-as-script quick checks
if __name__ == "__main__":
print("Variables available:", list(__all_vars__.keys()))
print("Matrix row 1:", matrix[1])
print("Counter value:", ctr.value)
Resources
Official Spyder documentation: Spyder Documentation
VS Code
Features
VS Code provides a flexible, extensible interface that is well suited for editing, debugging, and remote development. Below are the main panes, useful workflows, and configuration tips to get the most out of VS Code when working locally or connected to an HPC resource.
Main panes and useful panels
- Explorer (left) 📁
File tree and quick file operations.
Right-click to open terminals, reveal in OS file manager, or run commands.
- Editor (center) ✍️
Supports tabs, split editors, and editor groups.
Rich file-type support (syntax, linting, formatting).
Notebook editor for .ipynb files when Jupyter extension is installed.
- Side bar (left) 🔧
Run & Debug, Source Control, Extensions, Search, Remote Explorer.
- Panel (bottom) 🖥️
Integrated Terminal(s): run jobs, activate envs, and submit sbatch scripts.
Output, Problems, Debug Console, and Tests.
- Status bar (bottom) ℹ️
Shows current interpreter, Git branch, line endings, and active extensions.
Click the interpreter area to switch Python interpreters or kernels.
Quick Tips
Use the Command Palette (
Ctrl+Shift+P/F1) to find commands quickly (interpreter selection, remote commands, setting server install path).Open the integrated terminal (``Ctrl+` ``) to run modules, activate conda/venv, or submit batch jobs.
Use the Python: Select Interpreter command to point VS Code at the exact Python executable on the remote host (useful after module load or activate).
For notebooks, choose “Existing Jupyter Server” to connect VS Code to a Jupyter server running on a compute node.
Split the editor (
Ctrl+\) to compare files or keep docs and code side-by-side.
Recommended extensions
Python : language server, linting, testing, formatter hooks.
Jupyter : native notebook support and remote server attachment.
Remote-SSH : connect to HPC login nodes and work against remote files.
Keyboard shortcuts
Ctrl+P: Quick file openCtrl+Shift+P: Command Palette``Ctrl+` `` : Toggle integrated terminal
Ctrl+Shift+D: Open Run and DebugF5: Start debuggingCtrl+K Z: Zen Mode (distraction-free)
Configuring for remote development
Remote server install path:
change Remote.SSH: Server Install Pathto keep .vscode-server and extensions in a project directory with adequate space.Interpreter path: after loading modules or activating an environment on the remote host, run
Python: Select Interpreterand paste the full path fromwhich python.Port forwarding: when attaching to remote Jupyter or web UIs, check the Remote Explorer → Ports or Terminal → Ports view. VS Code can forward ports automatically.
Keep heavy extensions minimal on remote servers to save space and startup time. Prefer installing large language servers locally where possible.
Manage Extensions 🔧
By default, VSCode server installs all extensions in your home directory on the remote server. Which is not recommended as home directories have limited space. You can change this behavior by changing the remote server install path to a project folder with sufficient space.
Go to Command Palette
Ctrl+Shift+PorF1. Search forRemote-SSH: Settingsand then go toRemote.SSH: Server Install Path.Add
Itemas remote host (say, rackham.uppmax.uu.se) andValueas project folder in which you want to install all your data and extensions (say/proj/uppmax202x-x-xx/nobackup) (without a trailing slash /).
⚠️ If you already had your vscode-server running and storing extensions in home directory. Make sure to kill the server by selecting Remote-SSH: KIll VS Code Server on Host on Command Palette and deleting the .vscode-server directory in your home folder.
Install Extensions 📦
You can sync all your local VSCode extensions to the remote server after you are connected with VSCode server on HPC resource by searching for
Remote: Install Local ExtensionsinSSH: rackham.uppmax.uu.se(or other HPC name) in Command Palette.You can alternatively, go to Extensions tab and select each individually.
Selecting Kernels 🧠
Establish an SSH connection to the login node of the HPC resource using VSCode remote-SSH extension as described in previous session.
You may request an allocation on a compute node BUT VSCode server does not connect to it automatically and your code will still be executed on login node.
Load the correct module (or virtual env) on HPC resource that contains the interpreter you want on your VSCode. For example in case you need ML packages and python interpreter on Rackham/Snowy, do module load python_ML_packages. Check the file path for python interpreter by checking
which pythonand copy this path. Go to Command Palette Ctrl+Shift+P or F1 on your local VSCode. Search for “interpreter” for python, then paste the path of your interpreter/kernel.venv or conda environments are also visible on VSCode when you select interpreter/kernel for python or jupyter server.
NOTE: Fetching python interpreters from a compute node may or may not work depending on the HPC resource. Develop your code on login node and run it on compute nodes using sbatch scripts.
For Jupyter Notebooks (and a much safer option) 🧪
You need to start the server on the HPC resource first, preferably on a compute node.
Copy the jupyter server URL which goes something like
http://s193.uppmax.uu.se:8888/tree?token=xxx(where s193 is Snowy node. Other HPCs will have similar URLs), click onSelect Kernelon VSCode and selectExisting Jupyter Server. Past the URL here and confirm your choice.This only works if you have the jupyter extension installed on your local VSCode.
The application will automatically perform port forwarding to your local machine from the compute nodes over certain ports. Check the Terminal->Ports tab to see the correct url to open in your browser.
NOTE: Selecting kernels/interpreter does not work currently on HPC2N.
Resources
Exercises
Exercise
Try running a Scipy and a Pytorch example in your favorite IDE.
Create a Virtual env using your faviroute package manager and install the packages.
For an extra challenge: Run the same code in .ipynb format in your IDE. This requires you to install jupyter notebook in your virtual environment.
Solving linear system of equations and optimization task using Scipy
Install Scipy for the following example.
import numpy as np
from scipy.linalg import solve
from scipy.optimize import minimize
# Test 1: Solve a linear system of equations
# Ax = b
A = np.array([[3, 1], [1, 2]])
b = np.array([9, 8])
# Solve for x
x = solve(A, b)
print("Solution to the linear system Ax = b:")
print(x)
# Test 2: Minimize a simple quadratic function
# f(x) = (x - 3)^2
def quadratic_function(x):
return (x - 3) ** 2
# Initial guess
x0 = [0]
# Minimize the function
result = minimize(quadratic_function, x0)
print("\nOptimization result:")
print("Minimum value of f(x):", result.fun)
print("Value of x at minimum:", result.x)
Loading transformers model with pytorch backend and performing tokenization
Install transformers[torch] for the following example. Can be performed either on GPU or CPU node.
from transformers import AutoTokenizer, AutoModel
import torch
# Check if GPU is available
if torch.cuda.is_available():
device = torch.device("cuda")
print("GPU is available. Using:", torch.cuda.get_device_name(0))
else:
device = torch.device("cpu")
print("GPU is not available. Using CPU.")
# Load a pre-trained tokenizer and model
model_name = "bert-base-uncased"
tokenizer = AutoTokenizer.from_pretrained(model_name)
model = AutoModel.from_pretrained(model_name)
# Move the model to GPU (if available)
model = model.to(device)
# Check where the model is loaded
print(f"Model is loaded on: {device}")
# Tokenize a sample text
text = "Transformers library is amazing!"
inputs = tokenizer(text, return_tensors="pt").to(device) # Move inputs to GPU if available
# Perform a forward pass to ensure everything works
outputs = model(**inputs)
# Detokenize the input IDs back to text
detokenized_text = tokenizer.decode(inputs["input_ids"][0], skip_special_tokens=True)
print("Detokenized text:", detokenized_text)
- Learning outcomes:
How to use IDE on any system
How to run code on IDE
How to install and manage extensions on remote VSCode server