More about reproducibility
Versioning systems
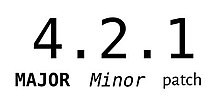
Semantic (SemVer)
Given a version number MAJOR.MINOR.PATCH, increment the:
MAJOR version when you make incompatible API changes
MINOR version when you add functionality in a backwards compatible manner
PATCH version when you make backwards compatible bug fixes Additional labels for pre-release and build metadata are available as extensions to the MAJOR.MINOR.PATCH format.
<https://nehckl0.medium.com/semver-and-calver-2-popular-software-versioning-schemes-96be80efe36
Calender (CalVer)
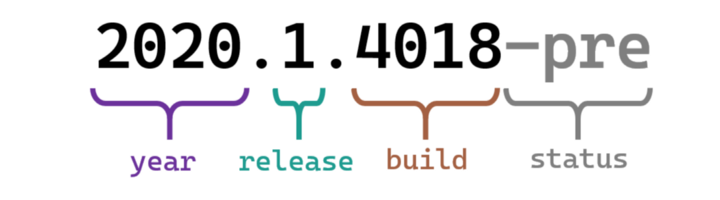
<https://nehckl0.medium.com/semver-and-calver-2-popular-software-versioning-schemes-96be80efe36
Recording dependencies
Reproducibility: We can control our code but how can we control dependencies?
10-year challenge: Try to build/run your own code that you have created 10 (or less) years ago. Will your code from today work in 5 years if you don’t change it?
Dependency hell: Different codes on the same environment can have conflicting dependencies.
Conda, Anaconda, pip, Virtualenv, Pipenv, pyenv, Poetry, requirements.txt …
These tools try to solve the following problems:
Defining a specific set of dependencies, possibly with well defined versions
Installing those dependencies mostly automatically
Recording the versions for all dependencies
Isolated environments
On your computer for projects so they can use different software.
Isolate environments on computers with many users (and allow self-installations)
Using different Python/R versions per project??
Provide tools and services to share packages
The tools
Python
PyPI -pip freeze > requirements.txt
Conda: any language, also compiled code and libraries.
conda-forge is a GitHub organization containing repositories of conda recipes.
Export the requirements into requirements.txt with conda list –export > requirements.txt.
Export the full environment using conda env export > environment.yml, and compare the .yml file format to the .txt file format.
Virtualenv
Pipenv
Poetry
Pyenv
Mamba (faster conda)
R
Packrat, jetpack, rsuite, renv, automagic, deplearning, devtools
C/C+
CMake
Conan
Conda
Fortran
Fortran package manager
Julia
Pkg.jl
designed around using isolated environments with independent sets of packages. Environments can either be local to a particular project or shared and selected by name.
Course advertisement
Recording computational steps
Questions
How can we create a reproducible workflow?
When to use scientific workflow management systems.
Objectives
Discuss pros/cons of GUI vs. manual steps vs. scripted vs. workflow tools.
4 ways (from CodeRefinery)
Graphical user interface (GUI)
Imagine we have programmed a GUI with a nice interface with icons where you can select scripts and input files by clicking:
Click on counting script
Select book txt file
Give a name for the dat file
Click on a run symbol
Click on plotting script
Select book dat file
Give a name for the image file
Click on a run symbol
…
Go to next book …
Click on counting script
Select book txt file
…
Disclaimer: not all GUIs behave this way - there exist very good GUI solutions which enable reproducibility and automation.
Solution 2: Manual steps
It is not too much work for four files:
$ python source/wordcount.py data/abyss.txt processed_data/abyss.dat
$ python source/plotcount.py processed_data/abyss.dat processed_data/abyss.png
$ python source/wordcount.py data/isles.txt processed_data/isles.dat
$ python source/plotcount.py processed_data/isles.dat processed_data/isles.png
$ python source/wordcount.py data/last.txt processed_data/last.dat
$ python source/plotcount.py processed_data/last.dat processed_data/last.png
$ python source/wordcount.py data/sierra.txt processed_data/sierra.dat
$ python source/plotcount.py processed_data/sierra.dat processed_data/sierra.png
$ python source/zipf_test.py processed_data/abyss.dat processed_data/isles.dat processed_data/last.dat processed_data/sierra.dat
This is imperative style: first do this, then to that, then do that, finally do …
Solution 3: Script
Let’s express it more compactly with a shell script (Bash). Let’s call it script.sh:
#!/usr/bin/env bash
# loop over all books
for title in abyss isles last sierra; do
python source/wordcount.py data/${title}.txt processed_data/${title}.dat
python source/plotcount.py processed_data/${title}.dat processed_data/${title}.png
done
# this could be done using variables but nevermind
python source/zipf_test.py processed_data/abyss.dat processed_data/isles.dat processed_data/last.dat processed_data/sierra.dat
We can run it with:
bash script.sh
Solution 4: Using Snakemake or other workflow manager
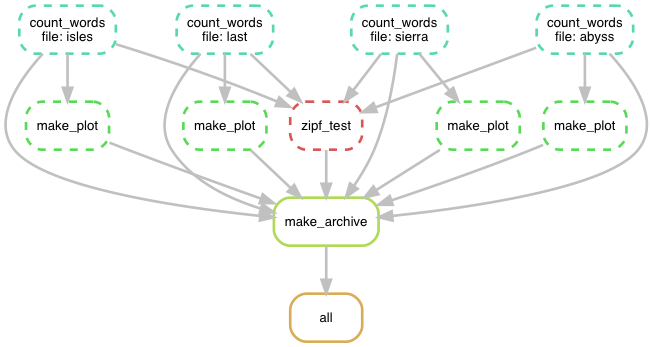
Isolated tracks of work.
Discussion
Discuss the pros and cons of these different approaches. Which are reproducible? Which scale to hundreds of books and which can it be automated?
Solution
GUIs may or may not be reproducible.
Some GUIs can be automated, many cannot.
Typing the same series of commands for 100 similar inputs is tedious and error prone.
Imperative scripts are reproducible and great for automation.
Declarative workflows such as Snakemake are great for longer multi-step analyses.
Declarative workflows are often easy to parallelize without you changing anything.
With declarative workflows it is no problem to add/change things late in the project.
Interesting modern alternative to Make/Snakemake: https://taskfile.dev
Many specialized frameworks exist.
Containers
Popular container implementations:
Docker
Singularity (popular on high-performance computing systems)
Apptainer (popular on high-performance computing systems, fork of Singularity)
podman
They are to some extent interoperable:
podman is very close to Docker
Docker images can be converted to Singularity/Apptainer images
Singularity Python can convert Dockerfiles to Singularity definition files
https://coderefinery.github.io/reproducible-research/environments
Reproducible publications
Git (overleaf, authorea), hackmd, manuscripts.io, google docs
Scholarly output reproducible: rrtools, jupyter, binder, research compendia
Reprohack
Keypoints
Preserve the steps for re-generating published results.
Hundreds of workflow management tools exist.
Snakemake is a comparatively simple and lightweight option to create transferable and scalable data analyses.
Sometimes a script is enough.