Parallel computing with Python
Questions
What is parallel computing?
What are the different parallelization mechanisms for Python?
How to implement parallel algorithms in Python code?
How to deploy threads and workers at our HPC centers?
Objectives
Learn general concepts for parallel computing
Gain knowledge on the tools for parallel programming in different languages
Familiarize with the tools to monitor the usage of resources
Prerequisites
Demo
If not already done so:
$ ml GCCcore/11.2.0 Python/3.9.6
$ virtualenv /proj/nobackup/<your-project-storage>/vpyenv-python-course
$ source /proj/nobackup/<your-project-storage>/vpyenv-python-course/bin/activate
For the
numbaexample install the corresponding module:
$ pip install numba
For the
mpi4pyexample add the following modules:
$ ml GCC/11.2.0 OpenMPI/4.1.1
$ pip install mpi4py
For the
f2pyexample, this command should be available on the terminal whennumpyis installed:
$ pip install numpy
For the Julia example we will need PyJulia:
$ ml Julia/1.9.3-linux-x86_64
$ pip install julia
Start Python on the command line and type:
>>> import julia
>>> julia.install()
Quit Python, you should be ready to go!
If not already done so:
$ module load python/3.9.5
$ python -m venv --system-site-packages /proj/naiss202X-XY-XYZ/nobackup/<user>/venv-python-course
Activate it if needed (is the name shown in the prompt)
$ source /proj/naiss202X-XY-XYZ/nobackup/<user>/venv-python-course/bin/activate
For the
numbaexample install the corresponding module:
$ python -m pip install numba
For the
mpi4pyexample add the following modules:
$ ml gcc/9.3.0 openmpi/3.1.5
$ python -m pip install mpi4py
For the Julia example we will need PyJulia:
$ ml julia/1.7.2
$ python -m pip install julia
Start Python on the command line and type:
>>> import julia
>>> julia.install()
Quit Python, you should be ready to go!
These guidelines are working for Tetralith:
$ ml buildtool-easybuild/4.8.0-hpce082752a2 GCCcore/11.3.0 Python/3.10.4
$ ml GCC/11.3.0 OpenMPI/4.1.4
$ python -m /path-to-your-project/venv vpyenv-python-course
$ source /path-to-your-project/vpyenv-python-course/bin/activate
For the
mpi4pyexample add the following modules:
$ pip install mpi4py
For the
numbaexample install the corresponding module:
$ pip install numba
For the Julia example we will need PyJulia:
$ ml julia/1.9.4-bdist
$ pip install JuliaCall
Start Julia on the command line and add the following package:
pkg> add PythonCall
In Python there are different schemes that can be used to parallelize your code. We will only take a look at some of these schemes that illustrate the general concepts of parallel computing. The aim of this lecture is to learn how to run parallel codes in Python rather than learning to write those codes.
The workhorse for this section will be a 2D integration example:
\[\int^{\pi}_{0}\int^{\pi}_{0}\sin(x+y)dxdy = 0\]
One way to perform the integration is by creating a grid in the x and y directions.
More specifically, one divides the integration range in both directions into n bins. A
serial code (without optimization) can be seen in the following code block.
integration2d_serial.pyimport math import sys from time import perf_counter # grid size n = 10000 def integration2d_serial(n): global integral; # interval size (same for X and Y) h = math.pi / float(n) # cummulative variable mysum = 0.0 # regular integration in the X axis for i in range(n): x = h * (i + 0.5) # regular integration in the Y axis for j in range(n): y = h * (j + 0.5) mysum += math.sin(x + y) integral = h**2 * mysum if __name__ == "__main__": starttime = perf_counter() integration2d_serial(n) endtime = perf_counter() print("Integral value is %e, Error is %e" % (integral, abs(integral - 0.0))) print("Time spent: %.2f sec" % (endtime-starttime))
We can run this code on the terminal as follows (similarly at both HPC2N and UPPMAX):
Warning
Although this works on the terminal, having many users doing computations at the same time for this course, could create delays for other users
$ python integration2d_serial.py
Integral value is -7.117752e-17, Error is 7.117752e-17
Time spent: 20.39 sec
Because of that, we can use for short-time jobs the following command:
$ srun -A <your-projec-id> -n 1 -t 00:10:00 python integration2d_serial.py
Integral value is -7.117752e-17, Error is 7.117752e-17
Time spent: 20.39 sec
where srun has the flags that are used in a standard batch file.
Note that outputs can be different, when timing a code a more realistic approach would be to run it several times to get statistics.
One of the crucial steps upon parallelizing a code is identifying its bottlenecks. In the present case, we notice that the most expensive part in this code is the double for loop.
Serial optimizations
Just before we jump into a parallelization project, Python offers some options to make
serial code faster. For instance, the Numba module can assist you to obtain a
compiled-quality function with minimal efforts. This can be achieved with the njit()
decorator:
integration2d_serial_numba.pyfrom numba import njit import math import sys from time import perf_counter # grid size n = 10000 def integration2d_serial(n): # interval size (same for X and Y) h = math.pi / float(n) # cummulative variable mysum = 0.0 # regular integration in the X axis for i in range(n): x = h * (i + 0.5) # regular integration in the Y axis for j in range(n): y = h * (j + 0.5) mysum += math.sin(x + y) integral = h**2 * mysum return integral if __name__ == "__main__": starttime = perf_counter() integral = njit(integration2d_serial)(n) endtime = perf_counter() print("Integral value is %e, Error is %e" % (integral, abs(integral - 0.0))) print("Time spent: %.2f sec" % (endtime-starttime))
The execution time is now:
$ python integration2d_serial_numba.py
Integral value is -7.117752e-17, Error is 7.117752e-17
Time spent: 1.90 sec
Another option for making serial codes faster, and specially in the case of arithmetic intensive codes, is to write the most expensive parts of them in a compiled language such as Fortran or C/C++. In the next paragraphs we will show you how Fortran code for the 2D integration case can be called in Python.
We start by writing the expensive part of our Python code in a Fortran function in a file
called fortran_function.f90:
fortran_function.f90function integration2d_fortran(n) result(integral) implicit none integer, parameter :: dp=selected_real_kind(15,9) real(kind=dp), parameter :: pi=3.14159265358979323_dp integer, intent(in) :: n real(kind=dp) :: integral integer :: i,j ! interval size real(kind=dp) :: h ! x and y variables real(kind=dp) :: x,y ! cummulative variable real(kind=dp) :: mysum h = pi/(1.0_dp * n) mysum = 0.0_dp ! regular integration in the X axis do i = 0, n-1 x = h * (i + 0.5_dp) ! regular integration in the Y axis do j = 0, n-1 y = h * (j + 0.5_dp) mysum = mysum + sin(x + y) enddo enddo integral = h*h*mysum end function integration2d_fortran
Then, we need to compile this code and generate the Python module
(myfunction):
Warning
For UPPMAX you may have to change gcc version like:
$ ml gcc/10.3.0
Then continue…
$ f2py -c -m myfunction fortran_function.f90
running build
running config_cc
...
this will produce the Python/C API myfunction.cpython-39-x86_64-linux-gnu.so, which
can be called in Python as a module:
call_fortran_code.pyfrom time import perf_counter import myfunction import numpy # grid size n = 10000 if __name__ == "__main__": starttime = perf_counter() integral = myfunction.integration2d_fortran(n) endtime = perf_counter() print("Integral value is %e, Error is %e" % (integral, abs(integral - 0.0))) print("Time spent: %.2f sec" % (endtime-starttime))
The execution time is considerably reduced:
$ python call_fortran_code.py
Integral value is -7.117752e-17, Error is 7.117752e-17
Time spent: 1.30 sec
Compilation of code can be tedious specially if you are in a developing phase of your code. As an alternative to improve the performance of expensive parts of your code (without using a compiled language) you can write these parts in Julia (which doesn’t require compilation) and then calling Julia code in Python. For the workhorse integration case that we are using, the Julia code can look like this:
julia_function.jlfunction integration2d_julia(n::Int) # interval size h = π/n # cummulative variable mysum = 0.0 # regular integration in the X axis for i in 0:n-1 x = h*(i+0.5) # regular integration in the Y axis for j in 0:n-1 y = h*(j + 0.5) mysum = mysum + sin(x+y) end end return mysum*h*h end
A caller script for Julia would be,
call_julia_code.pyfrom time import perf_counter import julia from julia import Main Main.include('julia_function.jl') # grid size n = 10000 if __name__ == "__main__": starttime = perf_counter() integral = Main.integration2d_julia(n) endtime = perf_counter() print("Integral value is %e, Error is %e" % (integral, abs(integral - 0.0))) print("Time spent: %.2f sec" % (endtime-starttime))from time import perf_counter from juliacall import Main as julia # Include the Julia script julia.include("julia_function.jl") # grid size n = 10000 if __name__ == "__main__": starttime = perf_counter() # Call the function defined in the julia script integral = julia.integration2d_julia(n) # function takes arguments endtime = perf_counter() print("Integral value is %e, Error is %e" % (integral, abs(integral - 0.0))) print("Time spent: %.2f sec" % (endtime-starttime))
Timing in this case is similar to the Fortran serial case,
$ python call_julia_code.py
Integral value is -7.117752e-17, Error is 7.117752e-17
Time spent: 1.29 sec
If even with the previous (and possibly others from your own) serial optimizations your code doesn’t achieve the expected performance, you may start looking for some parallelization scheme. Here, we describe the most common schemes.
Threads
In a threaded parallelization scheme, the workers (threads) share a global memory address space. The threading module is built into Python so you don’t have to installed it. By using this module, one can create several threads to do some work in parallel (in principle). For jobs dealing with files I/O one can observe some speedup by using the threading module. However, for CPU intensive jobs one would see a decrease in performance w.r.t. the serial code. This is because Python uses the Global Interpreter Lock (GIL) which serializes the code when several threads are used.
In the following code we used the threading module to parallelize the 2D integration example.
Threads are created with the construct threading.Thread(target=function, args=()), where
target is the function that will be executed by each thread and args is a tuple containing the
arguments of that function. Threads are started with the start() method and when they finish
their job they are joined with the join() method,
integration2d_threading.pyimport threading import math import sys from time import perf_counter # grid size n = 10000 # number of threads numthreads = 4 # partial sum for each thread partial_integrals = [None]*numthreads def integration2d_threading(n,numthreads,threadindex): global partial_integrals; # interval size (same for X and Y) h = math.pi / float(n) # cummulative variable mysum = 0.0 # workload for each thread workload = n/numthreads # lower and upper integration limits for each thread begin = int(workload*threadindex) end = int(workload*(threadindex+1)) # regular integration in the X axis for i in range(begin,end): x = h * (i + 0.5) # regular integration in the Y axis for j in range(n): y = h * (j + 0.5) mysum += math.sin(x + y) partial_integrals[threadindex] = h**2 * mysum if __name__ == "__main__": starttime = perf_counter() # start the threads threads = [] for i in range(numthreads): t = threading.Thread(target=integration2d_threading, args=(n,numthreads,i)) threads.append(t) t.start() # waiting for the threads for t in threads: t.join() integral = sum(partial_integrals) endtime = perf_counter() print("Integral value is %e, Error is %e" % (integral, abs(integral - 0.0))) print("Time spent: %.2f sec" % (endtime-starttime))
Notice the output of running this code on the terminal:
$ python integration2d_threading.py
Integral value is 4.492851e-12, Error is 4.492851e-12
Time spent: 21.29 sec
Although we are distributing the work on 4 threads, the execution time is longer than in the serial code. This is due to the GIL mentioned above.
Implicit Threaded
Some libraries like OpenBLAS, LAPACK, and MKL provide an implicit threading mechanism. They
are used, for instance, by numpy module for computing linear algebra operations. You can obtain information
about the libraries that are available in numpy with numpy.show_config().
This can be useful at the moment of setting the number of threads as these libraries could
use different mechanisms for it, for the following example we will use the OpenMP
environment variables.
Consider the following code that computes the dot product of a matrix with itself:
dot.pyfrom time import perf_counter import numpy as np A = np.random.rand(3000,3000) starttime = perf_counter() B = np.dot(A,A) endtime = perf_counter() print("Time spent: %.2f sec" % (endtime-starttime))
the timing for running this code with 1 thread is:
$ export OMP_NUM_THREADS=1
$ python dot.py
Time spent: 1.14 sec
while running with 2 threads is:
$ export OMP_NUM_THREADS=2
$ python dot.py
Time spent: 0.60 sec
It is also possible to use efficient threads if you have blocks of code written in a compiled language. Here, we will see the case of the Fortran code written above where OpenMP threads are used. The parallelized code looks as follows:
fortran_function_openmp.f90function integration2d_fortran_openmp(n) result(integral) !$ use omp_lib implicit none integer, parameter :: dp=selected_real_kind(15,9) real(kind=dp), parameter :: pi=3.14159265358979323 integer, intent(in) :: n real(kind=dp) :: integral integer :: i,j ! interval size real(kind=dp) :: h ! x and y variables real(kind=dp) :: x,y ! cummulative variable real(kind=dp) :: mysum h = pi/(1.0_dp * n) mysum = 0.0_dp ! regular integration in the X axis !$omp parallel do reduction(+:mysum) private(x,y,j) do i = 0, n-1 x = h * (i + 0.5_dp) ! regular integration in the Y axis do j = 0, n-1 y = h * (j + 0.5_dp) mysum = mysum + sin(x + y) enddo enddo !$omp end parallel do integral = h*h*mysum end function integration2d_fortran_openmp
The way to compile this code differs to the one we saw before, now we will need the flags for OpenMP:
$ f2py -c --f90flags='-fopenmp' -lgomp -m myfunction_openmp fortran_function_openmp.f90
the generated module can be then loaded,
call_fortran_code_openmp.pyfrom time import perf_counter import myfunction_openmp import numpy # grid size n = 10000 if __name__ == "__main__": starttime = perf_counter() integral = myfunction_openmp.integration2d_fortran_openmp(n) endtime = perf_counter() print("Integral value is %e, Error is %e" % (integral, abs(integral - 0.0))) print("Time spent: %.2f sec" % (endtime-starttime))
the execution time by using 4 threads is:
$ export OMP_NUM_THREADS=4
$ python call_fortran_code_openmp.py
Integral value is 4.492945e-12, Error is 4.492945e-12
Time spent: 0.37 sec
More information about how OpenMP works can be found in the material of a previous OpenMP course offered by some of us.
Distributed
In the distributed parallelization scheme the workers (processes) can share some common memory but they can also exchange information by sending and receiving messages for instance.
integration2d_multiprocessing.pyimport multiprocessing from multiprocessing import Array import math import sys from time import perf_counter # grid size n = 10000 # number of processes numprocesses = 4 # partial sum for each thread partial_integrals = Array('d',[0]*numprocesses, lock=False) def integration2d_multiprocessing(n,numprocesses,processindex): global partial_integrals; # interval size (same for X and Y) h = math.pi / float(n) # cummulative variable mysum = 0.0 # workload for each process workload = n/numprocesses begin = int(workload*processindex) end = int(workload*(processindex+1)) # regular integration in the X axis for i in range(begin,end): x = h * (i + 0.5) # regular integration in the Y axis for j in range(n): y = h * (j + 0.5) mysum += math.sin(x + y) partial_integrals[processindex] = h**2 * mysum if __name__ == "__main__": starttime = perf_counter() processes = [] for i in range(numprocesses): p = multiprocessing.Process(target=integration2d_multiprocessing, args=(n,numprocesses,i)) processes.append(p) p.start() # waiting for the processes for p in processes: p.join() integral = sum(partial_integrals) endtime = perf_counter() print("Integral value is %e, Error is %e" % (integral, abs(integral - 0.0))) print("Time spent: %.2f sec" % (endtime-starttime))
In this case, the execution time is reduced:
$ python integration2d_multiprocessing.py
Integral value is 4.492851e-12, Error is 4.492851e-12
Time spent: 6.06 sec
MPI
More details for the MPI parallelization scheme in Python can be found in a previous MPI course offered by some of us.
integration2d_mpi.pyfrom mpi4py import MPI import math import sys from time import perf_counter # MPI communicator comm = MPI.COMM_WORLD # MPI size of communicator numprocs = comm.Get_size() # MPI rank of each process myrank = comm.Get_rank() # grid size n = 10000 def integration2d_mpi(n,numprocs,myrank): # interval size (same for X and Y) h = math.pi / float(n) # cummulative variable mysum = 0.0 # workload for each process workload = n/numprocs begin = int(workload*myrank) end = int(workload*(myrank+1)) # regular integration in the X axis for i in range(begin,end): x = h * (i + 0.5) # regular integration in the Y axis for j in range(n): y = h * (j + 0.5) mysum += math.sin(x + y) partial_integrals = h**2 * mysum return partial_integrals if __name__ == "__main__": starttime = perf_counter() p = integration2d_mpi(n,numprocs,myrank) # MPI reduction integral = comm.reduce(p, op=MPI.SUM, root=0) endtime = perf_counter() if myrank == 0: print("Integral value is %e, Error is %e" % (integral, abs(integral - 0.0))) print("Time spent: %.2f sec" % (endtime-starttime))
Execution of this code gives the following output:
$ mpirun -np 4 python integration2d_mpi.py
Integral value is 4.492851e-12, Error is 4.492851e-12
Time spent: 5.76 sec
For long jobs, one will need to run in batch mode. Here is an example of a batch script for this MPI example,
#!/bin/bash
#SBATCH -A hpc2n20XX-XYZ
#SBATCH -t 00:05:00 # wall time
#SBATCH -n 4
#SBATCH -o output_%j.out # output file
#SBATCH -e error_%j.err # error messages
ml purge > /dev/null 2>&1
ml GCCcore/11.2.0 Python/3.9.6
ml GCC/11.2.0 OpenMPI/4.1.1
#ml Julia/1.7.1-linux-x86_64 # if Julia is needed
source /proj/nobackup/<your-project-storage>/vpyenv-python-course/bin/activate
mpirun -np 4 python integration2d_mpi.py
#!/bin/bash -l
#SBATCH -A naiss202X-XY-XYZ
#SBATCH -t 00:05:00
#SBATCH -n 4
#SBATCH -o output_%j.out # output file
#SBATCH -e error_%j.err # error messages
ml python/3.9.5
ml gcc/9.3.0 openmpi/3.1.5
#ml julia/1.7.2 # if Julia is needed
source /proj/naiss202X-XY-XYZ/nobackup/<user>/venv-python-course/bin/activate
mpirun -np 4 python integration2d_mpi.py
#!/bin/bash -l
#SBATCH -A naiss202X-XY-XYZ
#SBATCH -t 00:05:00
#SBATCH -n 4
#SBATCH -o output_%j.out # output file
#SBATCH -e error_%j.err # error messages
ml buildtool-easybuild/4.8.0-hpce082752a2 GCCcore/11.3.0 Python/3.10.4
ml GCC/11.3.0 OpenMPI/4.1.4
#ml julia/1.9.4-bdist # if Julia is needed
source /path-to-your-project/vpyenv-python-course/bin/activate
mpirun -np 4 python integration2d_mpi.py
Monitoring resources’ usage
Monitoring the resources that a certain job uses is important specially when this job is expected to run on many CPUs and/or GPUs. It could happen, for instance, that an incorrect module is loaded or the command for running on many CPUs is not the proper one and our job runs in serial mode while we allocated possibly many CPUs/GPUs. For this reason, there are several tools available in our centers to monitor the performance of running jobs.
HPC2N
On a Kebnekaise terminal, you can type the command:
$ job-usage job_ID
where job_ID is the number obtained when you submit your job with the sbatch
command. This will give you a URL that you can copy and then paste in your local
browser. The results can be seen in a graphical manner a couple of minutes after the
job starts running, here there is one example of how this looks like:
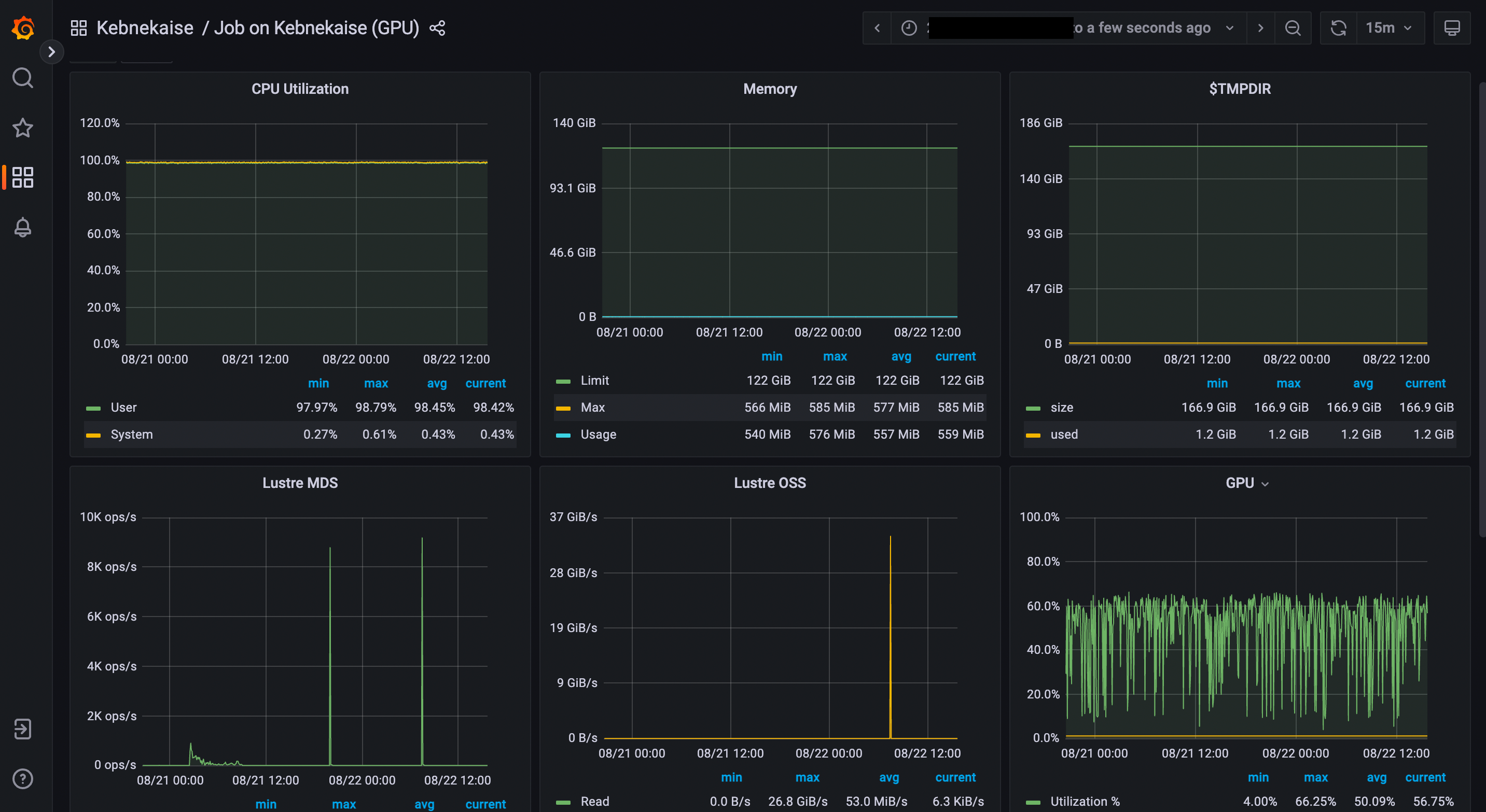
The resources used by a job can be monitored in your local browser. For this job, we can notice that 100% of the requested CPU and 60% of the GPU resources are being used.
Exercises
Running a parallel code efficiently
In this exercise we will run a parallelized code that performs a 2D integration:
\[\int^{\pi}_{0}\int^{\pi}_{0}\sin(x+y)dxdy = 0\]
One way to perform the integration is by creating a grid in the x and y directions.
More specifically, one divides the integration range in both directions into n bins.
Here is a parallel code using the multiprocessing module in Python (call it
integration2d_multiprocessing.py):
integration2d_multiprocessing.py
import multiprocessing
from multiprocessing import Array
import math
import sys
from time import perf_counter
# grid size
n = 5000
# number of processes
numprocesses = *FIXME*
# partial sum for each thread
partial_integrals = Array('d',[0]*numprocesses, lock=False)
# Implementation of the 2D integration function (non-optimal implementation)
def integration2d_multiprocessing(n,numprocesses,processindex):
global partial_integrals;
# interval size (same for X and Y)
h = math.pi / float(n)
# cummulative variable
mysum = 0.0
# workload for each process
workload = n/numprocesses
begin = int(workload*processindex)
end = int(workload*(processindex+1))
# regular integration in the X axis
for i in range(begin,end):
x = h * (i + 0.5)
# regular integration in the Y axis
for j in range(n):
y = h * (j + 0.5)
mysum += math.sin(x + y)
partial_integrals[processindex] = h**2 * mysum
if __name__ == "__main__":
starttime = perf_counter()
processes = []
for i in range(numprocesses):
p = multiprocessing.Process(target=integration2d_multiprocessing, args=(n,numprocesses,i))
processes.append(p)
p.start()
# waiting for the processes
for p in processes:
p.join()
integral = sum(partial_integrals)
endtime = perf_counter()
print("Integral value is %e, Error is %e" % (integral, abs(integral - 0.0)))
print("Time spent: %.2f sec" % (endtime-starttime))
Run the code with the following batch script.
job.sh
#!/bin/bash -l
#SBATCH -A naiss202X-XY-XYZ # your project_ID
#SBATCH -J job-serial # name of the job
#SBATCH -n *FIXME* # nr. tasks/coresw
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
# Load any modules you need, here for Python 3.11.8 and compatible SciPy-bundle
module load python/3.11.8
python integration2d_multiprocessing.py
#!/bin/bash
#SBATCH -A hpc2n202X-XYZ # your project_ID
#SBATCH -J job-serial # name of the job
#SBATCH -n *FIXME* # nr. tasks
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
# Do a purge and load any modules you need, here for Python
ml purge > /dev/null 2>&1
ml GCCcore/11.2.0 Python/3.9.6
python integration2d_multiprocessing.py
#!/bin/bash
#SBATCH -A lu202X-XX-XX # your project_ID
#SBATCH -J job-serial # name of the job
#SBATCH -n *FIXME* # nr. tasks
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
# reservation (optional)
#SBATCH --reservation=RPJM-course*FIXME*
# Do a purge and load any modules you need, here for Python
ml purge > /dev/null 2>&1
ml GCCcore/12.3.0 Python/3.11.3
python integration2d_multiprocessing.py
#!/bin/bash -l
#SBATCH -A naiss202X-XY-XYZ # your project_ID
#SBATCH -J job-serial # name of the job
#SBATCH -n *FIXME* # nr. tasks/coresw
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
# Load any modules you need, here for Python 3.11.8 and compatible SciPy-bundle
ml buildtool-easybuild/4.8.0-hpce082752a2 GCCcore/11.3.0 Python/3.10.4
python integration2d_multiprocessing.py
Try different number of cores for this batch script (FIXME string) using the sequence: 1,2,4,8,12, and 14. Note: this number should match the number of processes (also a FIXME string) in the Python script. Collect the timings that are printed out in the job.*.out. According to these execution times what would be the number of cores that gives the optimal (fastest) simulation?
Challenge: Increase the grid size (n) to 15000 and submit the batch job with 4 workers (in the
Python script) and request 5 cores in the batch script. Monitor the usage of resources
with tools available at your center, for instance top (UPPMAX) or
job-usage (HPC2N).
Parallelizing a for loop workflow (Advanced)
Create a Data Frame containing two features, one called ID which has integer values from 1 to 10000, and the other called Value that contains 10000 integers starting from 3 and goes in steps of 2 (3, 5, 7, …). The following codes contain parallelized workflows whose goal is to compute the average of the whole feature Value using some number of workers. Substitute the FIXME strings in the following codes to perform the tasks given in the comments.
The main idea for all languages is to divide the workload across all workers. You can run the codes as suggested for each language.
Pandas is available in the following combo ml GCC/12.3.0 SciPy-bundle/2023.07 (HPC2N) and
ml python/3.11.8 (UPPMAX). Call the script script-df.py.
import pandas as pd
import multiprocessing
# Create a DataFrame with two sets of values ID and Value
data_df = pd.DataFrame({
'ID': range(1, 10001),
'Value': range(3, 20002, 2) # Generate 10000 odd numbers starting from 3
})
# Define a function to calculate the sum of a vector
def calculate_sum(values):
total_sum = *FIXME*(values)
return *FIXME*
# Split the 'Value' column into chunks of size 1000
chunk_size = *FIXME*
value_chunks = [data_df['Value'][*FIXME*:*FIXME*] for i in range(0, len(data_df['*FIXME*']), *FIXME*)]
# Create a Pool of 4 worker processes, this is required by multiprocessing
pool = multiprocessing.Pool(processes=*FIXME*)
# Map the calculate_sum function to each chunk of data in parallel
results = pool.map(*FIXME: function*, *FIXME: chunk size*)
# Close the pool to free up resources, if the pool won't be used further
pool.close()
# Combine the partial results to get the total sum
total_sum = sum(results)
# Compute the mean by dividing the total sum by the total length of the column 'Value'
mean_value = *FIXME* / len(data_df['*FIXME*'])
# Print the mean value
print(mean_value)
Run the code with the batch script:
#!/bin/bash -l
#SBATCH -A naiss2024-22-107 # your project_ID
#SBATCH -J job-parallel # name of the job
#SBATCH -n 4 # nr. tasks/coresw
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
# Load any modules you need, here for Python 3.11.8 and compatible SciPy-bundle
module load python/3.11.8
python script-df.py
#!/bin/bash
#SBATCH -A hpc2n202x-XXX # your project_ID
#SBATCH -J job-parallel # name of the job
#SBATCH -n 4 # nr. tasks
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
# Load any modules you need, here for Python 3.11.3 and compatible SciPy-bundle
module load GCC/12.3.0 Python/3.11.3 SciPy-bundle/2023.07
python script-df.py
#!/bin/bash
#SBATCH -A lu202X-XX-XX # your project_ID
#SBATCH -J job-parallel # name of the job
#SBATCH -n 4 # nr. tasks
#SBATCH --time=00:20:00 # requested time
#SBATCH --error=job.%J.err # error file
#SBATCH --output=job.%J.out # output file
#SBATCH --reservation=RPJM-course*FIXME* # reservation (optional)
# Purge and load any modules you need, here for Python & SciPy-bundle
ml purge
ml GCCcore/12.3.0 Python/3.11.3 SciPy-bundle/2023.07
python script-df.py
Solution
import pandas as pd
import multiprocessing
# Create a DataFrame with two sets of values ID and Value
data_df = pd.DataFrame({
'ID': range(1, 10001),
'Value': range(3, 20002, 2) # Generate 10000 odd numbers starting from 3
})
# Define a function to calculate the sum of a vector
def calculate_sum(values):
total_sum = sum(values)
return total_sum
# Split the 'Value' column into chunks
chunk_size = 1000
value_chunks = [data_df['Value'][i:i+chunk_size] for i in range(0, len(data_df['Value']), chunk_size)]
# Create a Pool of 4 worker processes, this is required by multiprocessing
pool = multiprocessing.Pool(processes=4)
# Map the calculate_sum function to each chunk of data in parallel
results = pool.map(calculate_sum, value_chunks)
# Close the pool to free up resources, if the pool won't be used further
pool.close()
# Combine the partial results to get the total sum
total_sum = sum(results)
# Compute the mean by dividing the total sum by the total length of the column 'Value'
mean_value = total_sum / len(data_df['Value'])
# Print the mean value
print(mean_value)
See also
The book High Performance Python is a good resource for ways of speeding up Python code.
Keypoints
You deploy cores and nodes via SLURM, either in interactive mode or batch
In Python, threads, distributed and MPI parallelization can be used.