Big data with Python
“Learning outcomes”
Learners can
allocate resources sufficient to data size
decide on useful file formats
use data-chunking as technique
To cover
File format
- Methods
RAM allocation
chunking
Files formats
Types of scientific data
Bit and Byte
The smallest building block of storage in the computer is a bit, which stores either a 0 or 1.
Normally a number of 8 bits are combined in a group to make a byte.
One byte (8 bits) can represent/hold at most 2^8 distinct values. Organising bytes in different ways can represent different types of information, i.e. data.
Numerical data
Different numerical data types (e.g. integer and floating-point numbers) can be represented by bytes. The more bytes we use for each value, the larger is the range or precision we get, but more bytes require more memory.
- DataTypes
16-bit: short (integer
32-bit: Int (integer)
32-bit: Single (floating point
64-bit: Double (floating point
Text data
- DataTypes
8-bit: char
more
When it comes to text data, the simplest character encoding is ASCII (American Standard Code for Information Interchange) and was the most common character encodings until 2008 when UTF-8 took over.
The original ASCII uses only 7 bits for representing each character and therefore encodes only 128 specified characters. Later it became common to use an 8-bit byte to store each character in memory, providing an extended ASCII.
As computers became more powerful and the need for including more characters from other languages like Chinese, Greek and Arabic became more pressing, UTF-8 became the most common encoding. UTF-8 uses a minimum of one byte and up to four bytes per character.
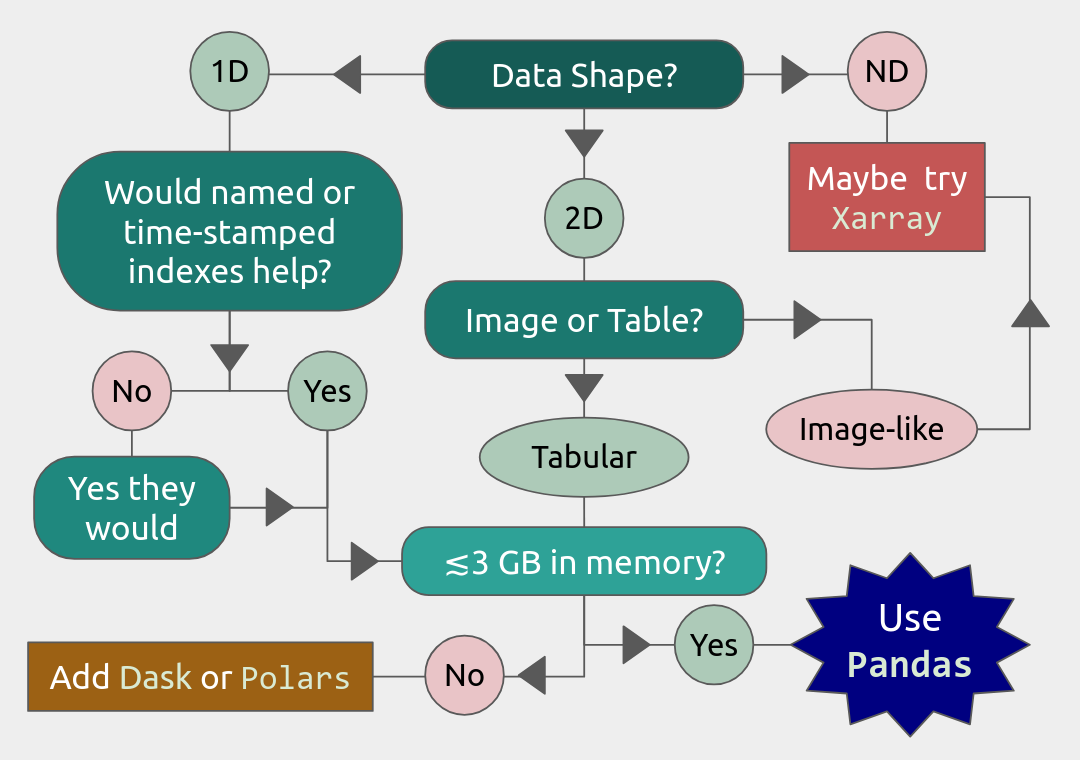
Data and storage format
In real scientific applications, data is complex and structured and usually contains both numerical and text data. Here we list a few of the data and file storage formats commonly used.
Tabular data
A very common type of data is “tabular data”.
Tabular data is structured into rows and columns.
Each column usually has a name and a specific data type while each row is a distinct sample which provides data according to each column (including missing values).
The simplest and most common way to save tabular data is via the so-called CSV (comma-separated values) file.
Gridded data
Gridded data is another very common data type in which numerical data is normally saved in a multi-dimensional rectangular grid. Most probably it is saved in one of the following formats:
more
Hierarchical Data Format (HDF5) - Container for many arrays
Network Common Data Form (NetCDF) - Container for many arrays which conform to the NetCDF data model
Zarr - New cloud-optimized format for array storage
CSV (comma-separated values)
Best use cases: Sharing data. Small data. Data that needs to be human-readable.
Key features
Type: Text format Packages needed: NumPy, Pandas Space efficiency: Bad Good for sharing/archival: Yes
more
- Tidy data:
Speed: Bad Ease of use: Great
- Array data:
Speed: Bad Ease of use: Ok for one or two dimensional data. Bad for anything higher.
HDF5 (Hierarchical Data Format version 5)
HDF5 is a high performance storage format for storing large amounts of data in multiple datasets in a single file.
It is especially popular in fields where you need to store big multidimensional arrays such as physical sciences.
Best use cases: Working with big datasets in array data format.
Key features
Type: Binary format
Packages needed: Pandas, PyTables, h5py
Space efficiency: Good for numeric data.
Good for sharing/archival: Yes, if datasets are named well.
more
- Tidy data:
Speed: Ok
Ease of use: Good
- Array data:
Speed: Great
Ease of use: Good
NETCDF4 (Network Common Data Form version 4)
NetCDF4 is a data format that uses HDF5 as its file format, but it has standardized structure of datasets and metadata related to these datasets.
This makes it possible to be read from various different programs.
Best use cases: Working with big datasets in array data format. Especially useful if the dataset contains spatial or temporal dimensions. Archiving or sharing those datasets.
Key features
Type: Binary format
Packages needed: Pandas, netCDF4/h5netcdf, xarray
Space efficiency: Good for numeric data.
Good for sharing/archival: Yes.
more
- Tidy data:
Speed: Ok
Ease of use: Good
- Array data:
Speed: Good
Ease of use: Great
NetCDF4 is by far the most common format for storing large data from big simulations in physical sciences.
The advantage of NetCDF4 compared to HDF5 is that one can easily add additional metadata, e.g. spatial dimensions (x, y, z) or timestamps (t) that tell where the grid-points are situated. As the format is standardized, many programs can use this metadata for visualization and further analysis.
XARRAY
Xarray is a Python package that builds on NumPy but adds labels to multi-dimensional arrays.
It also borrows heavily from the Pandas package for labelled tabular data and integrates tightly with dask for parallel computing.
Xarray is particularly tailored to working with NetCDF files.
- It reads and writes to NetCDF file using
open_dataset()functionopen_dataarray()functionto_netcdf()method.
Explore these in the exercise below!
See also
ENCCS course “HPDA-Python”: Scientific data
Aalto Scientific Computing course “Python for Scientific Computing”: Xarray
Allocating RAM
Important
Allocate many cores or a full node!
You do not have to explicitely run threads or other parallelism.
Note that shared memory among the cores works within node only
How much memory do I get per core?
Divide GB RAM of the booked node with number of cores.
- Example: 128 GB node with 20 cores
~6.4 GB per core
How much memory do I get with 5 cores?
Multiply the RAM per core with number of allocated cores..
- Example: 6.4 GB per core
~32 GB
Do you remember how to allocate several cores?
Slurm flag
-n <number of cores>
- Choose, if necessary a node with more RAM
See local HPC center documentation in how to do so!
Table of hardware
Technology |
Kebnekaise |
Rackham |
Snowy |
Bianca |
Cosmos |
Tetralith |
|---|---|---|---|---|---|---|
Cores/compute node |
28 (72 for largemem, 128/256 for AMD Zen3/Zen4) |
20 |
16 |
16 |
48 |
32 |
Memory/compute node |
128-3072 GB |
128-1024 GB |
128-4096 GB |
128-512 GB |
256-512 GB |
96-384 GB |
GPU |
NVidia V100, A100, A6000, L40s, H100, A40, AMD MI100 |
None |
NVidia T4 |
NVidia A100 |
NVidia A100 |
NVidia T4 |
Dask
Important
How to use more resources than available!!
Dask is very popular for data analysis and is used by a number of high-level Python libraries:
Dask-ML scales Scikit-Learn
Dask is composed of two parts:
Dynamic task scheduling optimized for computation. Similar to other workflow management systems, but optimized for interactive computational workloads.
“Big Data” collections like parallel arrays, dataframes, and lists that extend common interfaces like NumPy, Pandas, or Python iterators to larger-than-memory or distributed environments. These parallel collections run on top of dynamic task schedulers.
Dask Collections
Dask provides dynamic parallel task scheduling and three main high-level collections:
dask.array: Parallel NumPy arraysscales NumPy (see also xarray)
dask.dataframe: Parallel Pandas DataFramesscales Pandas workflows
dask.bag: Parallel Python Lists
Dask Arrays
A Dask array looks and feels a lot like a NumPy array.
- However, a Dask array uses the so-called “lazy” execution mode, which allows one to
build up complex, large calculations symbolically
before turning them over the scheduler for execution.
Dask divides arrays into many small pieces (chunks), as small as necessary to fit it into memory.
Operations are delayed (lazy computing) e.g. tasks are queue and no computation is performed until you actually ask values to be computed (for instance print mean values).
Then data is loaded into memory and computation proceeds in a streaming fashion, block-by-block.
Example from dask.org
# Arrays implement the Numpy API
import dask.array as da
x = da.random.random(size=(10000, 10000),
chunks=(1000, 1000))
x + x.T - x.mean(axis=0)
# It runs using multiple threads on your machine.
# It could also be distributed to multiple machines
Exercises
Load and run
Important
You should for this session load
ml GCC/12.3.0 Python/3.11.3 SciPy-bundle/2023.07 matplotlib/3.7.2 Tkinter/3.11.3
And install
dask&xarrayto~/.local/if you don’t already have it
pip install xarray dask
Important
You should for this session load
ml GCC/13.2.0 Python/3.11.5 SciPy-bundle/2023.11 matplotlib/3.8.2
And install
dask&xarrayto~/.local/if you don’t already have it
pip install xarray dask
Important
You should for this session load
module load python_ML_packages/3.11.8-cpu
And install
xarrayto~/.local/if you don’t already have it
pip install --user xarray
Important
You should for this session load
module load buildtool-easybuild/4.8.0-hpce082752a2 GCC/13.2.0 Python/3.11.5 SciPy-bundle/2023.11 JupyterLab/4.2.0
And install
dask&xarrayto~/.local/if you don’t already have it
pip install xarray dask
Note
You can do thes in the Python command line or in Jupyter.
Use Xarray to work with NetCDF files
This exercise is derived from Xarray Tutorials, which is distributed under an Apache-2.0 License.
First create an Xarray dataset:
import numpy as np
import xarray as xr
ds1 = xr.Dataset(
data_vars={
"a": (("x", "y"), np.random.randn(4, 2)),
"b": (("z", "x"), np.random.randn(6, 4)),
},
coords={
"x": np.arange(4),
"y": np.arange(-2, 0),
"z": np.arange(-3, 3),
},
)
ds2 = xr.Dataset(
data_vars={
"a": (("x", "y"), np.random.randn(7, 3)),
"b": (("z", "x"), np.random.randn(2, 7)),
},
coords={
"x": np.arange(6, 13),
"y": np.arange(3),
"z": np.arange(3, 5),
},
)
Then write the datasets to disk using to_netcdf() method:
ds1.to_netcdf("ds1.nc")
ds2.to_netcdf("ds2.nc")
You can read an individual file from disk by using open_dataset() method:
ds3 = xr.open_dataset("ds1.nc")
or using the load_dataset() method:
ds4 = xr.load_dataset('ds1.nc')
Tasks:
Explore the hierarchical structure of the
ds1andds2datasets in a Jupyter notebook by typing the variable names in a code cell and execute. Click the disk-looking objects on the right to expand the fields.Explore
ds3andds4datasets, and compare them withds1. What are the differences?
Chunk size
The following example calculate the mean value of a random generated array. Run the example and see the performance improvement by using dask.
import numpy as np
%%time
x = np.random.random((20000, 20000))
y = x.mean(axis=0)
import dask
import dask.array as da
%%time
x = da.random.random((20000, 20000), chunks=(1000, 1000))
y = x.mean(axis=0)
y.compute()
But what happens if we use different chunk sizes? Try out with different chunk sizes:
What happens if the dask chunks=(20000,20000)
What happens if the dask chunks=(250,250)
Choice of chunk size
The choice is problem dependent, but here are a few things to consider:
Each chunk of data should be small enough so that it fits comforably in each worker’s available memory. Chunk sizes between 10MB-1GB are common, depending on the availability of RAM. Dask will likely manipulate as many chunks in parallel on one machine as you have cores on that machine. So if you have a machine with 10 cores and you choose chunks in the 1GB range, Dask is likely to use at least 10 GB of memory. Additionally, there should be enough chunks available so that each worker always has something to work on.
On the otherhand, you also want to avoid chunk sizes that are too small as we see in the exercise. Every task comes with some overhead which is somewhere between 200us and 1ms. Very large graphs with millions of tasks will lead to overhead being in the range from minutes to hours which is not recommended.
Keypoints
Dask uses lazy execution
Only use Dask for processing very large amount of data
See also
Working with data
Tidy data
ENCCS
Dask for scalable analysis
Too be included in the future?