Packages
R packages
R packages is the main way of extending the functionallity of R and broadens the use of R to almost infinity!
Instead of writing code yourself there may be others that have done the same!
Many scientific tools are distributed as R packages, making it possible to run a script in the prompt and there define files to be analysed and arguments defining exactly what to do.
For more details about packages and in particular developing your own, see: R packages
Questions
What is an R package?
How do I find which packages and versions are available?
What to do if I need other packages?
Are there differences between HPC2N and UPPMAX?
Objectives
Understand the basics of what an R package is
Show how to check for R packages
show how to install own packages on the different clusters
R packages: A short Primer
What is a package, really?
An R package is essentially a contained folder and file structure containing R code (and possibly C/C++ or other code) and other files relevant for the package e.g. documentation(vignettes), licensing and configuration files. Let us look at a very simple example
$ git clone https://github.com/MatPiq/R_example.git
$ cd R_example
$ tree
.
├── DESCRIPTION
├── NAMESPACE
├── R
│ └── hello.R
├── man
│ └── hello.Rd
└── r_example.Rproj
Installing tree as non-root on Linux Ubuntu
If you are on a Linux Ubuntu system where tree is not installed, and you do not have root permissions, you can do this to install it in your own area
Create a directory (in your home folder) to install in:
Change to that directory:
cd ~/mytreeNow download tree:
apt download treeUnpack the files:
dpkg-deb -xv ./*deb ./You can use tree like this now, giving the full path:
~/mytree/usr/bin/treeNote: if you want to be able to use it with the command “tree” you could set an alias in your ~/.bashrc file and then
sourceit:echo 'alias tree="$HOME/mytree/usr/bin/tree"' >> ~/.bashrc source ~/.bashrc
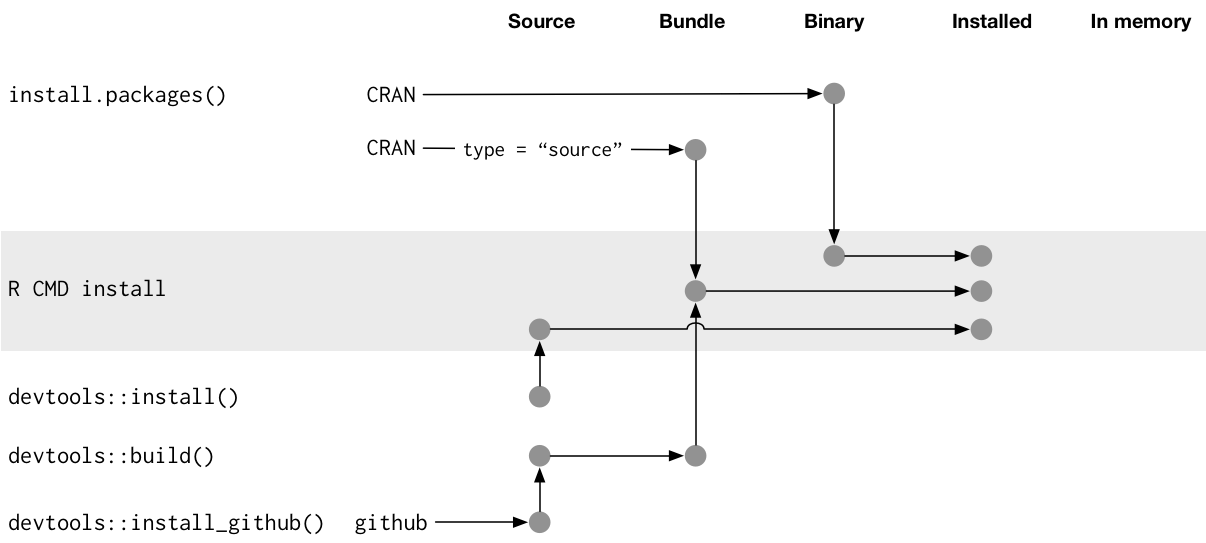
Package states
An R packages can exist in five possible states
Source: “source code” or “source files”. Development form.
Bundled: The source code compressed into a single file, usually tar.gz and sometimes referred to as “source tarballs”. Files in .Rbuildignore are excluded.
Binary: A compressed and pre-compiled version of a bundle built for a specific architecture. Usually how the package is provided by CRAN. Much faster than having to compile yourself and no need for dev/build tools.
Installed: A decompressed binary package located in a package _library_ (more on this later).
In-memory: When the installed package has been loaded from the library into memory, using require(pkg) or library(pkg).
Package libraries
In R, a library is a directory containing installed packages, sort of like a library for books. Unfortunately, in the R world, you will frequently encounter confused usage of the words “library” and “package”. It’s common for someone to refer to dplyr, for example, as a library when it is actually a package (Wickham & Hadley, 2023).
We might want to know where the R interpreter will be searching for
packages, i.e. where the libraries are located (could be several). The easiest
way to check is probably starting the interpreter and running the libPaths() function.
Load R, e.g. version 4.1.1 and start the Interpreter
$ ml R/4.1.1
$ R
Then check find the path of the library using the libPaths() function.
> .libPaths()
[1] "/sw/apps/R/4.1.1/rackham/lib64/R/library"
Load R, e.g. version 4.1.2 and start the Interpreter
$ ml GCC/11.2.0 OpenMPI/4.1.1 R/4.1.2
$ R
Then check find the path of the library using the libPaths() function.
> .libPaths()
[1] "/pfs/stor10/users/home/b/bbrydsoe/R-packages-4.1.2"
[2] "/cvmfs/ebsw.hpc2n.umu.se/amd64_ubuntu2004_bdw/software/R/4.1.2-foss-2021b/lib/R/library"
Preinstalled package libraries
Both UPPMAX and HPC2N offer a large amount of preinstalled packages.
HPC2N
On HPC2N most of these (around 750 packages) come with the
Rmodule and additional ones in theR-bundle-Bioconductor.NOTE: that on HPC2N, there are currently only two versions of the
R-bundle-Bioconductormodule, one that is compatible withR/4.0.0and one withR/4.1.2. Thus, if you need the extra packages included in theR-bundle-Bioconductormodule, you should use one of the compatible R versions. Usemodule spider <module>/<version>to check for prerequisites, as usual.
UPPMAX
On UPPMAX the module R_packages is a package library containing almost all packages in the CRAN and BioConductor repositories. As of 2023-10-11 there are a total of:
A total of 23476 R packages are installed
A total of 23535 packages are available in CRAN and BioConductor
19809 CRAN packages are installed, out of 19976 available
3544 BioConductor-specific packages are installed, out of 3559 available
121 other R packages are installed. These are not in CRAN/BioConductor, are only available in the CRAN/BioConductor archives, or are hosted on github, gitlab or elsewhere
There are many different ways to check if the package you are after is already installed - chances are it is! The simplest way is probably to simply try loading the package from within R
library(package-name)
Another option would be to create a dataframe of all the installed packages
ip <- as.data.frame(installed.packages()[,c(1,3:4)])
rownames(ip) <- NULL
ip <- ip[is.na(ip$Priority),1:2,drop=FALSE]
print(ip, row.names=FALSE)
However, this might not be so helpful unless you do additional filtering.
<br>
Another simple option is to grep the library directory. For example, both when loading R_packages at UPPMAX and R-bundle-Bioconductor at HPC2N the environment variable R_LIBS_SITE will be set to the path of the package
library.
Load R_packages
$ ml R_packages/4.1.1
Then grep for some package
$ ls -l $R_LIBS_SITE | grep glmnet
dr-xr-sr-x 9 douglas sw 4096 Sep 6 2021 EBglmnet
dr-xr-sr-x 11 douglas sw 4096 Nov 11 2021 glmnet
dr-xr-sr-x 8 douglas sw 4096 Sep 7 2021 glmnetcr
dr-xr-sr-x 7 douglas sw 4096 Sep 7 2021 glmnetUtils
Load R-bundle-Bioconductor
$ ml GCC/11.2.0 OpenMPI/4.1.1 R-bundle-Bioconductor/3.14-R-4.1.2
Check the R_LIBS_SITE environment variable
$ echo $R_LIBS_SITE
/hpc2n/eb/software/R-bundle-Bioconductor/3.14-foss-2021b-R-4.1.2:/hpc2n/eb/software/arrow-R/6.0.0.2-foss-2021b-R-4.1.2
Then grep for some package in the BioConductor package library
$ ls -l /hpc2n/eb/software/R-bundle-Bioconductor/3.14-foss-2021b-R-4.1.2 | grep RNA
drwxr-xr-x 9 easybuild easybuild 4096 Dec 30 2021 DeconRNASeq/
drwxr-xr-x 7 easybuild easybuild 4096 Dec 30 2021 RNASeqPower/
Installing your own packages
Sometimes you will need R packages that are not already installed. The solution to this is to install your own packages. These packages will usually come from CRAN (https://cran.r-project.org/) - the Comprehensive R Archive Network, or sometimes from other places, like GitHub or R-Forge
Here we will look at installing R packages with automatic download and with manual download. It is also possible to install from inside Rstudio.
Setup
We need to create a place for the own-installed packages to be and to tell R where to find them. The initial setup only needs to be done once, but separate package directories need to be created for each R version used.
R reads the $HOME/.Renviron file to setup its environment. It should be
created by R on first run, or you can create it with the command: touch
$HOME/.Renviron
NOTE: In this example we are going to assume you have chosen to place the R packages in a directory under your home directory, but in general it might be good to use the project storage for space reasons. As mentioned, you will need separate ones for each R version.
If you have not yet installed any packages to R yourself, the environment file should be empty and you can update it like this:
$ echo R_LIBS_USER="$HOME/R-packages-%V" > ~/.Renviron
Warning
If it is not empty, you can edit
$HOME/.Renvironwith your favorite editor so thatR_LIBS_USERcontains the path to your chosen directory for own-installed R packages.
It should look something like this when you are done:
$ R_LIBS_USER="/home/u/user/R-packages-%V"
NOTE Replace /home/u/user with the value of $HOME. Run echo $HOME to see its value.
NOTE The %V should be written as-is, it’s substituted at runtime with the active R version.
For each version of R you are using, create a directory matching the pattern
used in .Renviron to store your packages in. This example is shown for R
version 4.1.1:
$ mkdir -p $HOME/R-packages-4.1.1
Automatical download and install from CRAN
Note
You find a list of packages in CRAN (https://cran.r-project.org/) and a list of repos here: https://cran.r-project.org/mirrors.html
Please choose a location close to you when picking a repo.
$ R --quiet --no-save --no-restore -e "install.packages('<r-package>', repos='<repo>')"
install.packages('<r-package>', repos='<repo>')
In either case, the dependencies of the package will be downloaded and installed as well.
Example
In this example, we will install the R package stringr and use the
repository http://ftp.acc.umu.se/mirror/CRAN/
Note: You need to load R (and any prerequisites, and possibly R-bundle-Bioconductor if you need packages from that) before installing packages.
$ R --quiet --no-save --no-restore -e "install.packages('stringr', repos='http://ftp.acc.umu.se/mirror/CRAN/')"
install.packages('stringr', repos='http://ftp.acc.umu.se/mirror/CRAN/')
Automatic download and install from GitHub
If you want to install a package that is not on CRAN, but which do have a GitHub page, then there is an automatic way of installing, but you need to handle prerequsites yourself by installing those first. It can also be that the package is not in as finished a state as those on CRAN, so be careful.
To install packages from GitHub directly, from inside R, you first need to install the devtools package. Note that you only need to install this once.
This is how you install a package from GitHub, inside R:
install.packages("devtools") # ONLY ONCE devtools::install_github("DeveloperName/package")
Example
Type-Along
In this example we want to install the package quantstrat. It is not on CRAN, so let’s get it from the GitHub page for the project:
https://github.com/braverock/quantstrat
We also need to install devtools so we can install packages from GitHub. In
addition, quantstrat has some prerequisites, some on CRAN, some on GitHub,
so we need to install those as well.
install.packages("devtools") # ONLY ONCE
install.packages("FinancialInstrument")
install.packages("PerformanceAnalytics")
devtools::install_github("braverock/blotter")
devtools::install_github("braverock/quantstrat")
Manual download and install
If the package is not on CRAN or you want the development version, or you for other reason want to install a package you downloaded, then this is how to install from the command line:
$ R CMD INSTALL -l <path-to-R-package>/R-package.tar.gz
NOTE that if you install a package this way, you need to handle any dependencies yourself.
Note
Places to look for R packages
CRAN (https://cran.r-project.org/)
R-Forge (https://r-forge.r-project.org/)
Project’s own GitHub page
etc.
Keypoints
- You can check for installed packages
from inside R with
installed.packages()- from BASH shell with the
ml help R/<version>at UPPMAXml spider R/<version>at HPC2N
Installation of R packages can be done either from within R or from the command line (BASH shell)
CRAN is the recommended place to look for R-packages, but many packages can be found on GitHub and if you want the development version of a package you likely need to get it from GitHub or other place outside CRAN. You would then either download and install manually or install with something like devtools, from within R.
Install own packages on Bianca
If an R package is not not available on Bianca already (like Conda repositories) you may have to use the wharf to install the library/package
Typical workflow
Install on Rackham
Transfer to Wharf
Move package to local Bianca R package path
Test your installation
- Demo and exercise from our Bianca course:
Exercises
Install a package with automatic download
First do the setup of .Renviron and create the directory for installing R packages (Recommended load R version 4.1.1 on Rackham, 4.1.2 on Kebnekaise)
From the command line. Suggestion:
anomalizeFrom inside R. Suggestion: tidyr
Start R and see if the library can be loaded.
These are both on CRAN, and this way any dependencies will be installed as well.
Remember to pick a repo that is nearby, to install from: https://cran.r-project.org/mirrors.html
Solution for 4.1.1 on Rackham (change <user>)
$ echo R_LIBS_USER=\"$HOME/R-packages-%V\" > ~/.Renviron
R_LIBS_USER="/home/<user>/R-packages-%V"
$ mkdir -p $HOME/R-packages-4.1.1
Installing package “anomalize”. Using the repo http://ftp.acc.umu.se/mirror/CRAN/
$ R --quiet --no-save --no-restore -e "install.packages('anomalize', repo='http://ftp.acc.umu.se/mirror/CRAN/')"
This assumes you have already loaded the R module. If not, then do so first.
Installing package “tidyr”. Using the repo http://ftp.acc.umu.se/mirror/CRAN/
> install.packages('tidyr', repo='http://ftp.acc.umu.se/mirror/CRAN/')
$ R > library("anomalize") > library("tidyr")
“anomalize” outputs some text/advertisment when loaded. You can ignore this.